library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(aes(color = species)) +
theme_minimal()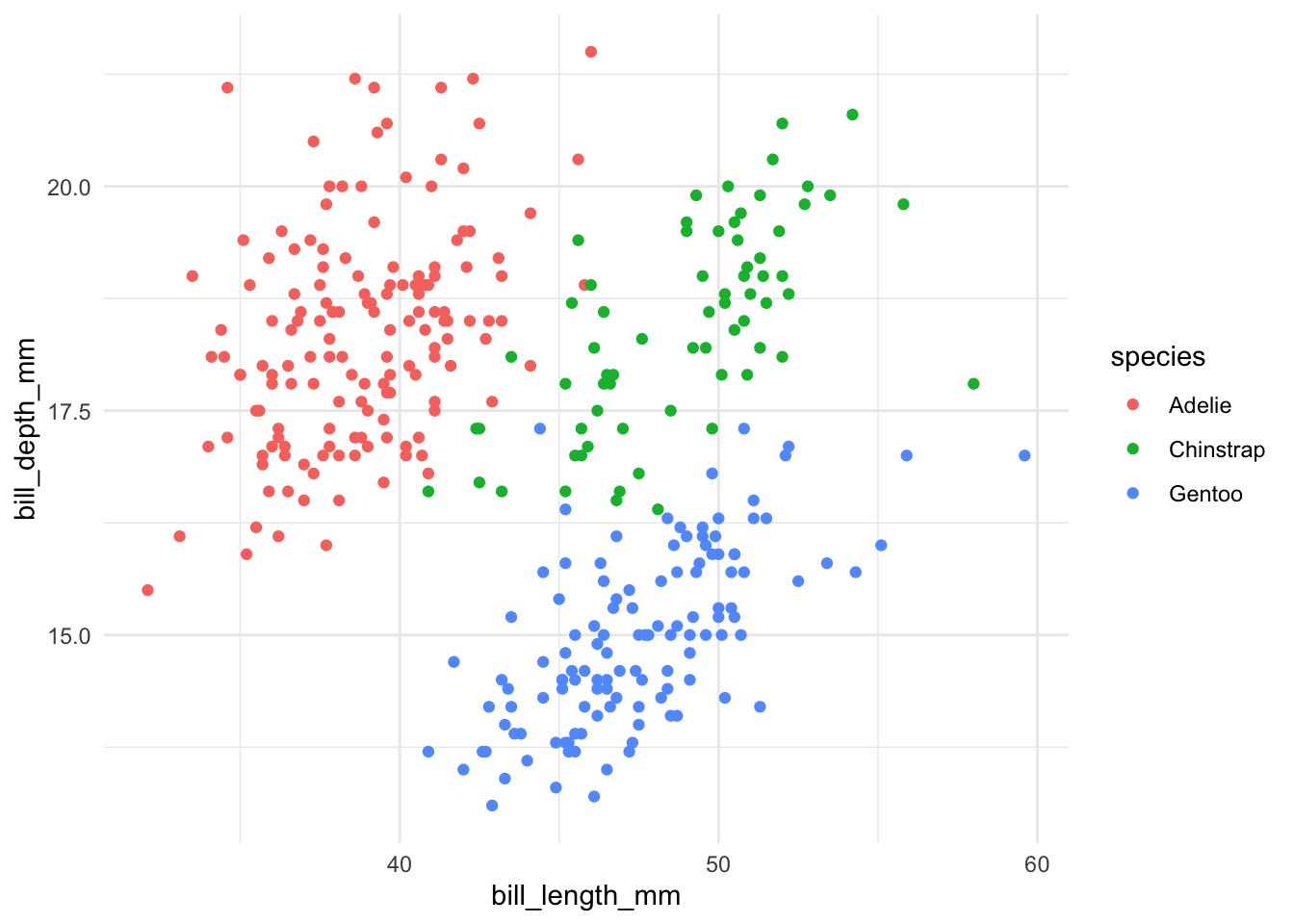
While many ggplot2 tutorials dive headfirst into creating various plot types (e.g., bar plots, line plots – the usual suspects). We’ll get to those, but first, let’s talk about what truly sets ggplot2 apart: its powerful customization. Although other plotting libraries might offer seemingly magical one-line solutions, these rarely survive contact with the harsh realities of academic publication and presentation. Effective visualizations require careful customization to address specific research questions. This guide covers key steps to create self-contained, informative, and aesthetically pleasing graphics—graphics that will make your readers (and reviewers) happy.
First, let’s rewind to where we left off in the previous chapter:
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(aes(color = species)) +
theme_minimal()
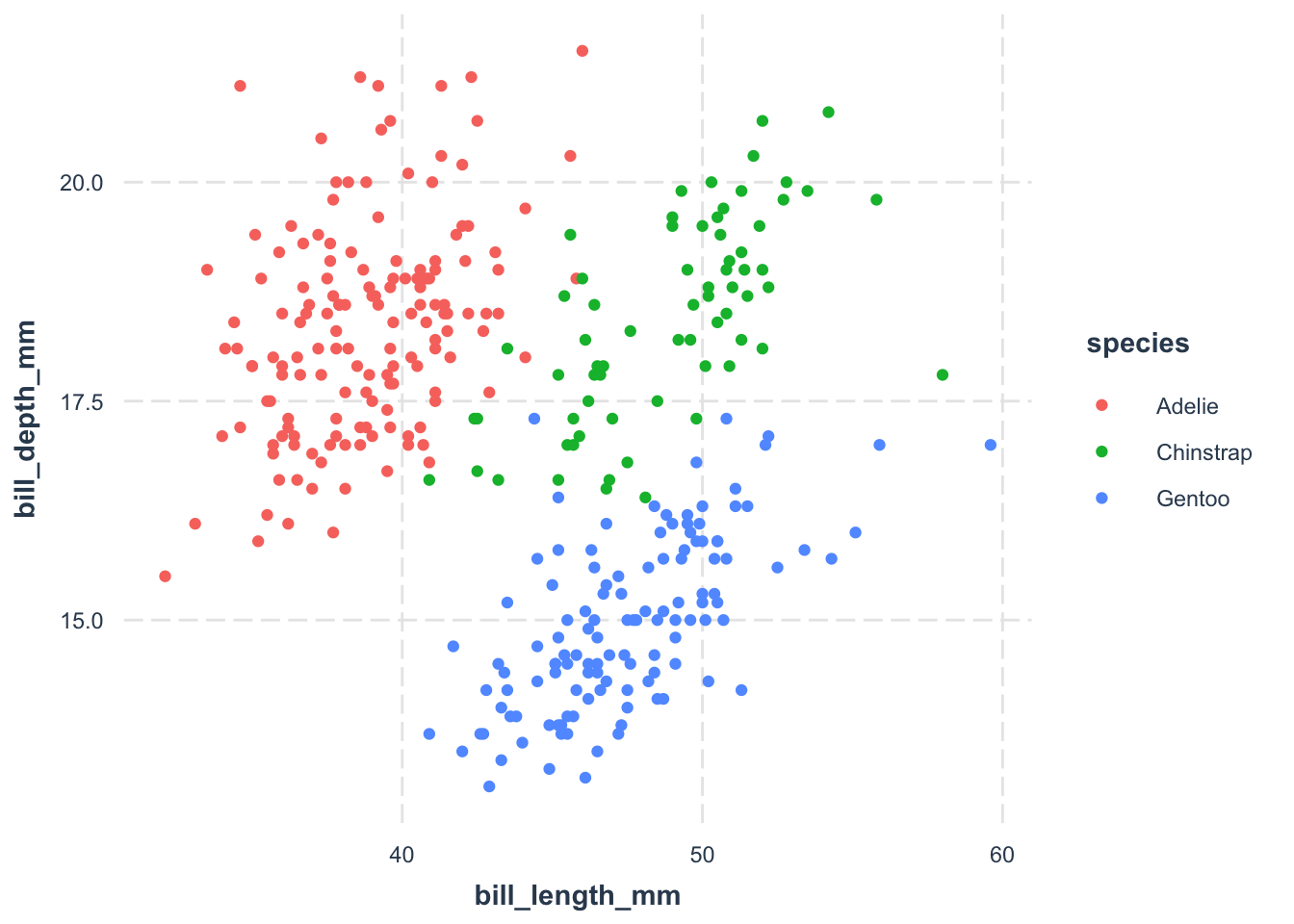
Remember, themes control the non-data elements of your plot. In the ggplot2 universe, the theme_*() functions offer opinionated customization options to tweak your plot’s appearance. Picking a theme you like can save you a boatload of time. Here are some choices I like:
I have a soft spot for theme_nice() from the jtools package. It’s clean, minimalistic, and easy on the eyes.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(aes(color = species)) +
jtools::theme_nice()theme_nice() function from the jtools package to apply the nice theme to the plot. If you have not installed jtools package yet, just run install.packages("jtools").
ggthemr offers a delightful collection of themes for ggplot2. It’s not on CRAN, so you’ll need to install it from GitHub.
pacman::p_load(devtools)
install_github('Mikata-Project/ggthemr')Here’s how to use the fresh theme in it, one of my favorites.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(aes(color = species)) +
ggthemr::ggthemr('fresh', set_theme = FALSE)$theme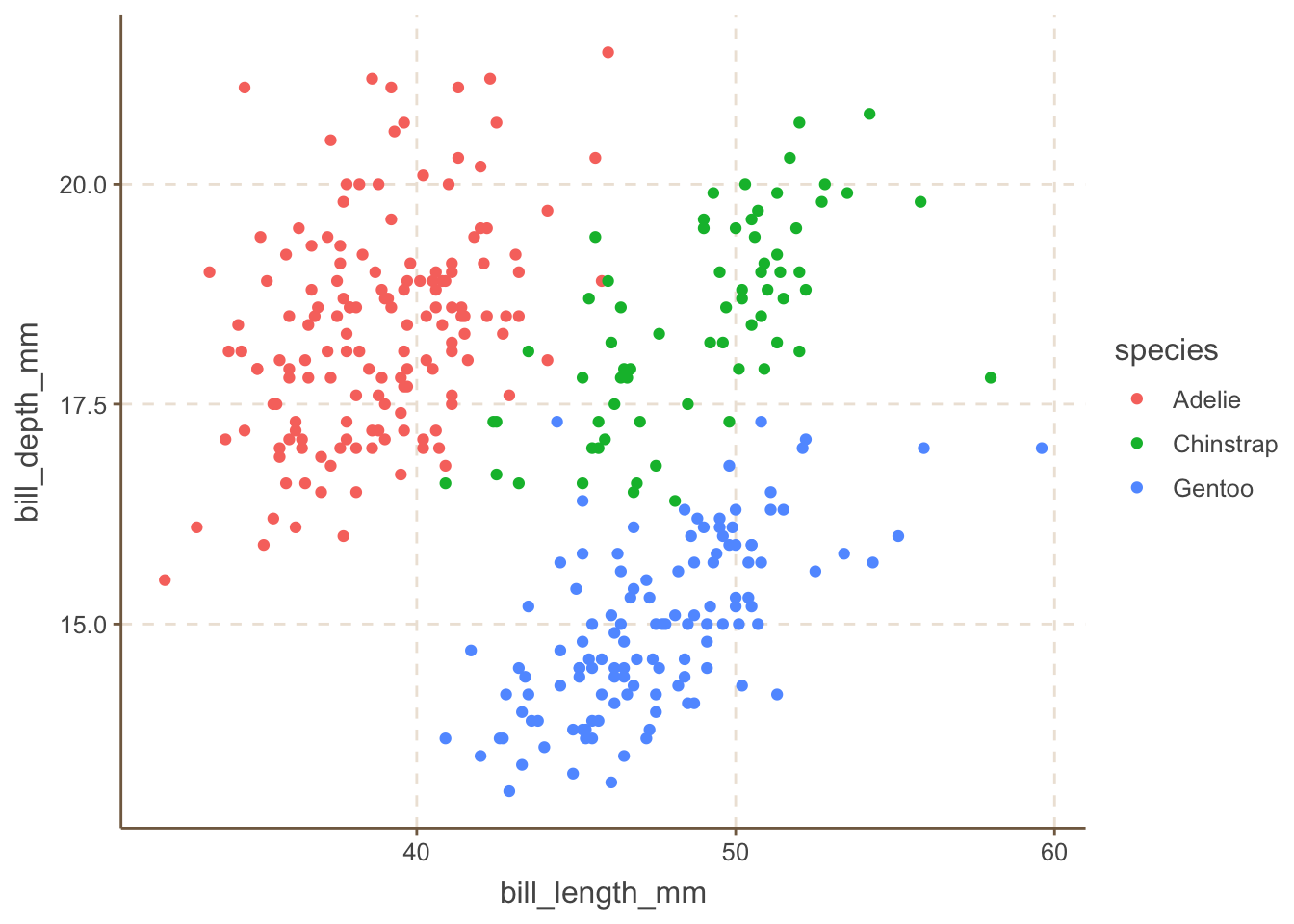
For more options, check out its GitHub page
Another contender is hrbrthemes, which also offers a suite of ggplot2 themes. I’m not its biggest fan, but it is quite popular.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(aes(color = species)) +
hrbrthemes::theme_ipsum()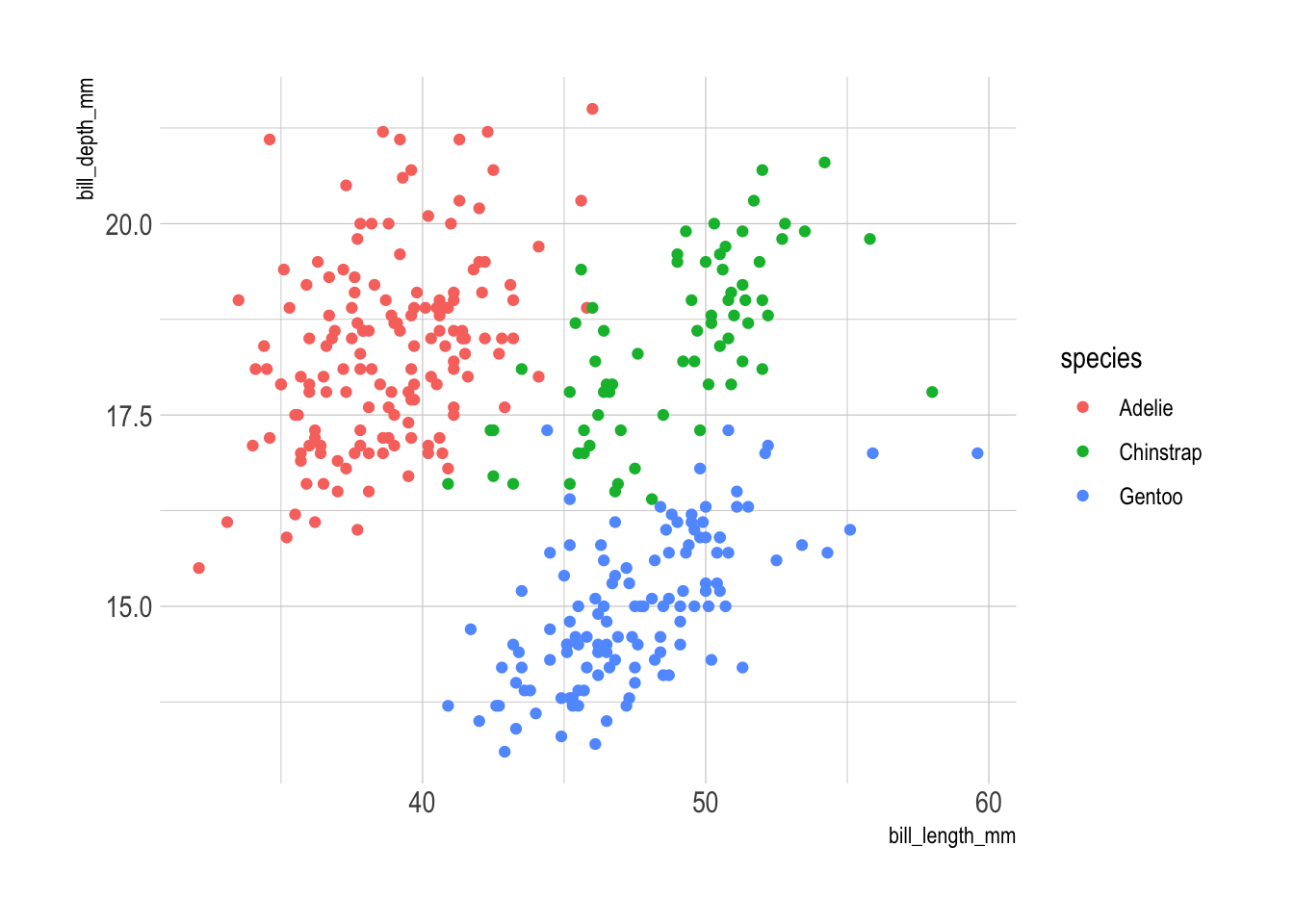
For those who prefer dark background, hrbrthemes has you covered:
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(aes(color = species)) +
hrbrthemes::theme_modern_rc()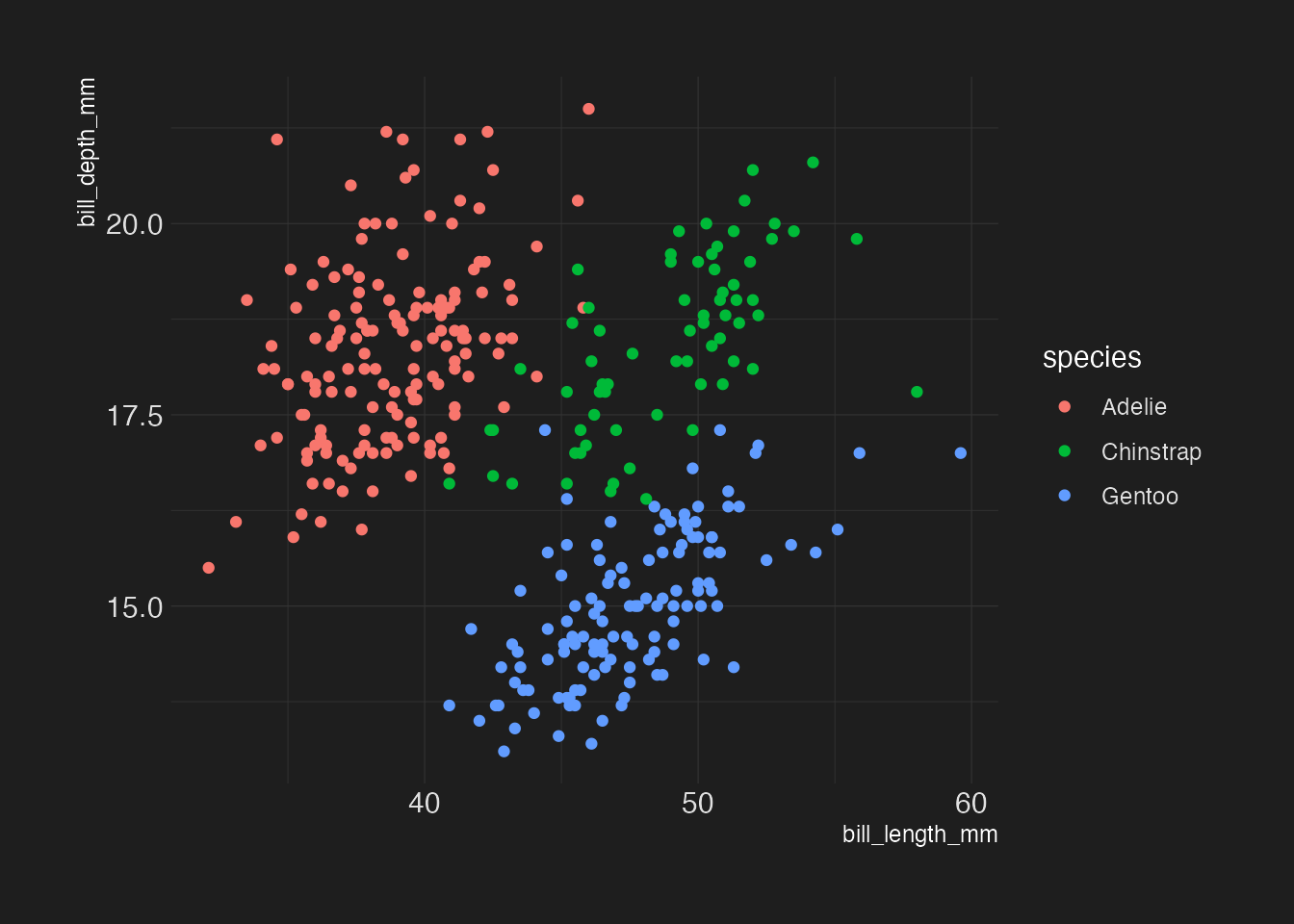
There are many other themes out there, so feel free to explore and find one that suits your style.
iris is a widely known dataset introduced by Ronald Fisher. It contains three plant species and four features measured for each sample. We will use this dataset to explore the association between Sepal.Width and Petal.Width as exercise throughout this Chapter. Try three different themes on the iris dataset:
As one example, you can use jtools::theme_nice() as the theme.
A common issue with scatter plots is that the boundary of points are unclear among different groups. We will use a new geometric object to lay the boundary of the points clear. We will use the package ggforce to add an ellipse around the points. As usually, if you have not installed the package yet, just run install.packages("ggforce").
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
ggforce::geom_mark_ellipse(
aes(fill = species),
alpha = 0.05,
color = 'transparent'
) +
geom_point(aes(color = species)) +
jtools::theme_nice()geom_mark_ellipse() function from the ggforce package to add an ellipse around the points.
fill = species argument.
color = 'transparent' argument.
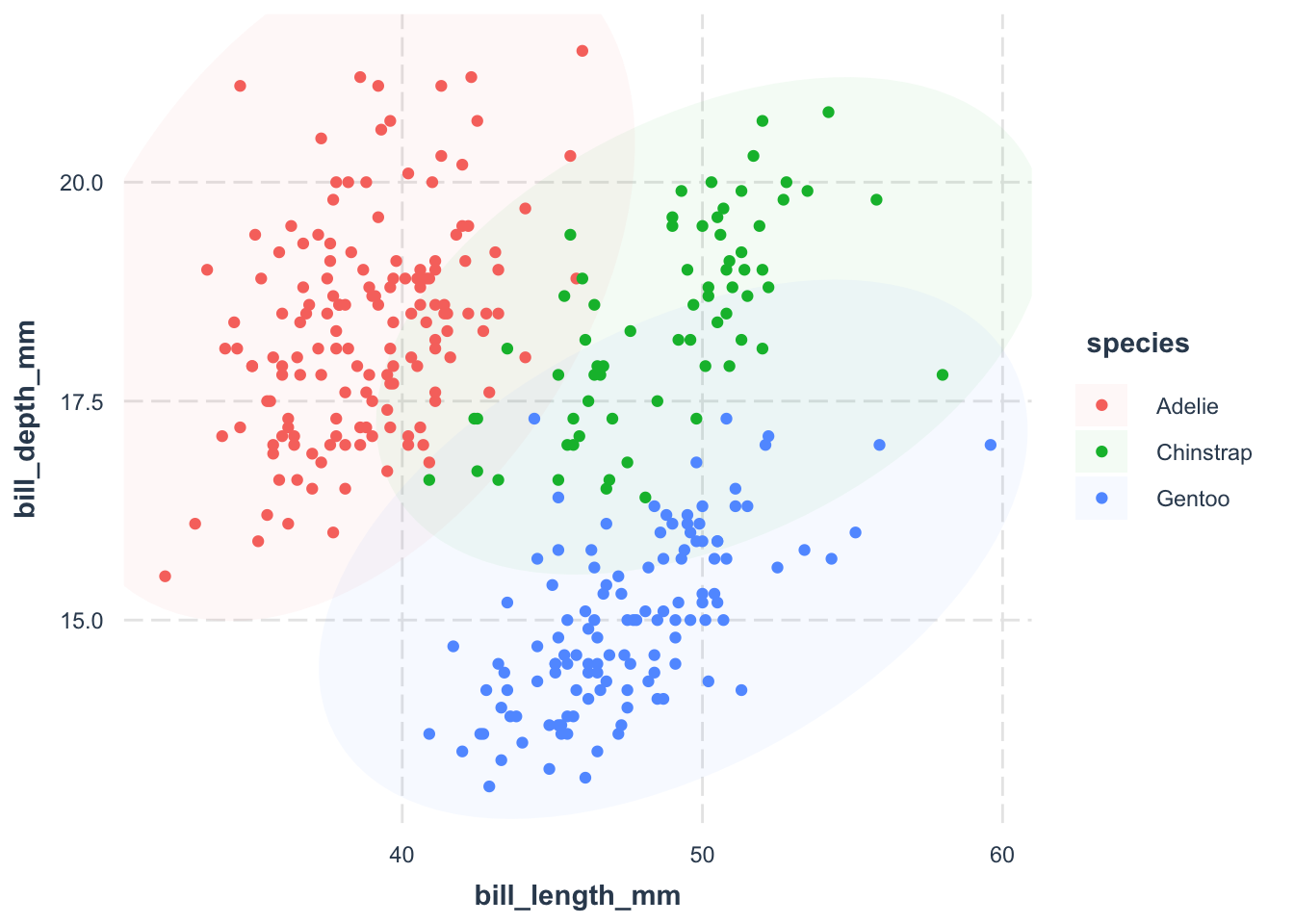
ggforce::geom_mark_ellipse() is a great way to show the distribution of points. However, it may not be the best choice for large datasets, especially when we want to show the density of points.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
ggdensity::geom_hdr(aes(fill = species)) +
# geom_point(shape = 21) +
jtools::theme_nice()geom_hdr() function from the ggdensity package to create a density plot. The fill = species argument specifies that we want to fill the density plot with the color of the species.
geom_point() function to add points to the density plot. The shape = 21 argument specifies that we want to use point shape 21, which is a circle with a border.
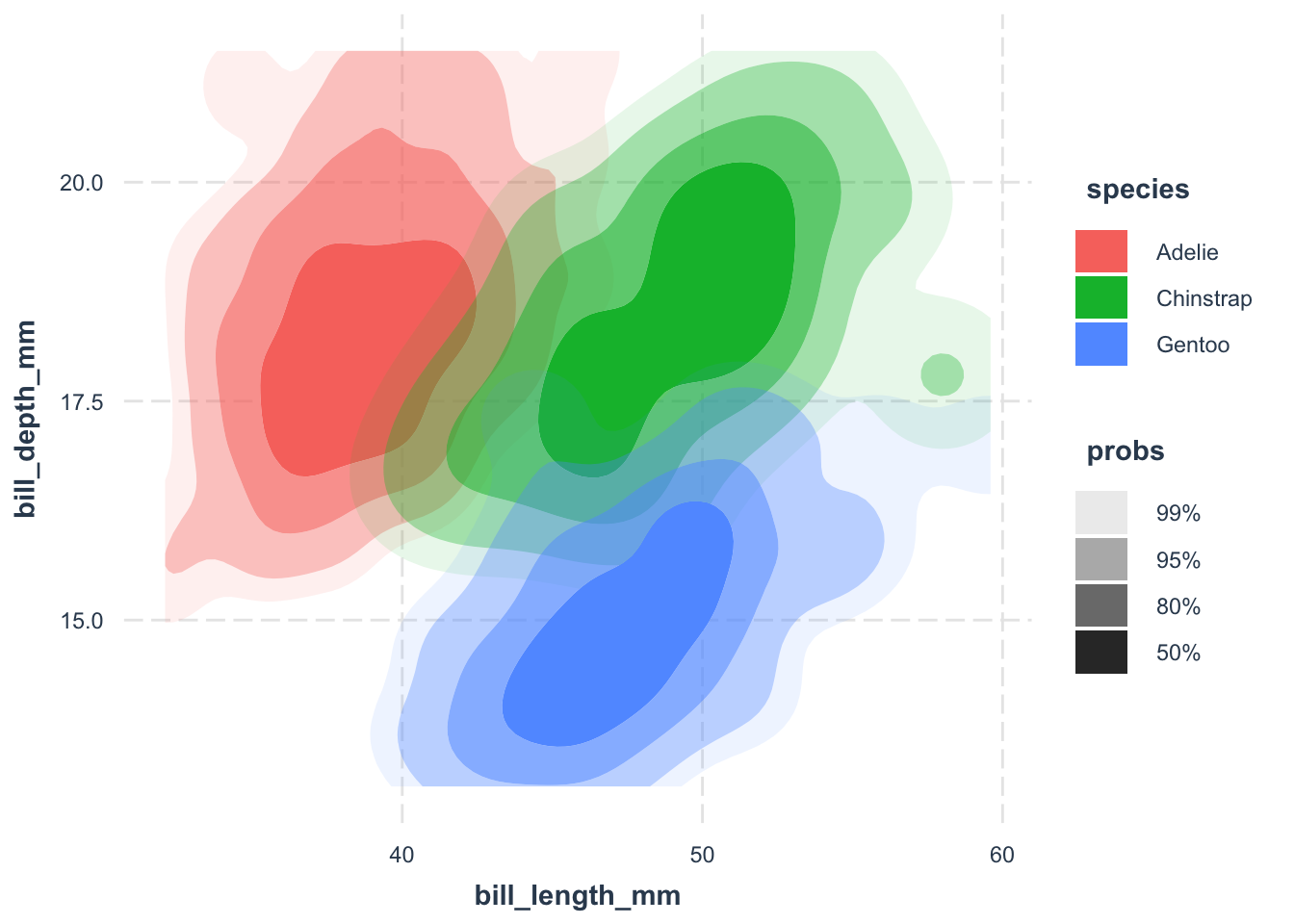
The default points are aesthetically not pleasing, at least to me. Customizing them can make your plot visually appealing. A common trick is to use point shape 21—a circle with a border. Fill the circle with the species color, add some transparency, and give it a white border. This way, overlapping points don’t turn into an unrecognizable blob.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
ggforce::geom_mark_ellipse(
aes(fill = species),
alpha = 0.05,
color = 'transparent'
) +
geom_point(
aes(fill = species),
color = "white",
shape = 21,
alpha = .6,
size = 3
) +
jtools::theme_nice()fill = species argument.
color = "white" argument.
shape = 21 argument.
alpha = .6 argument.
size = 3 argument.
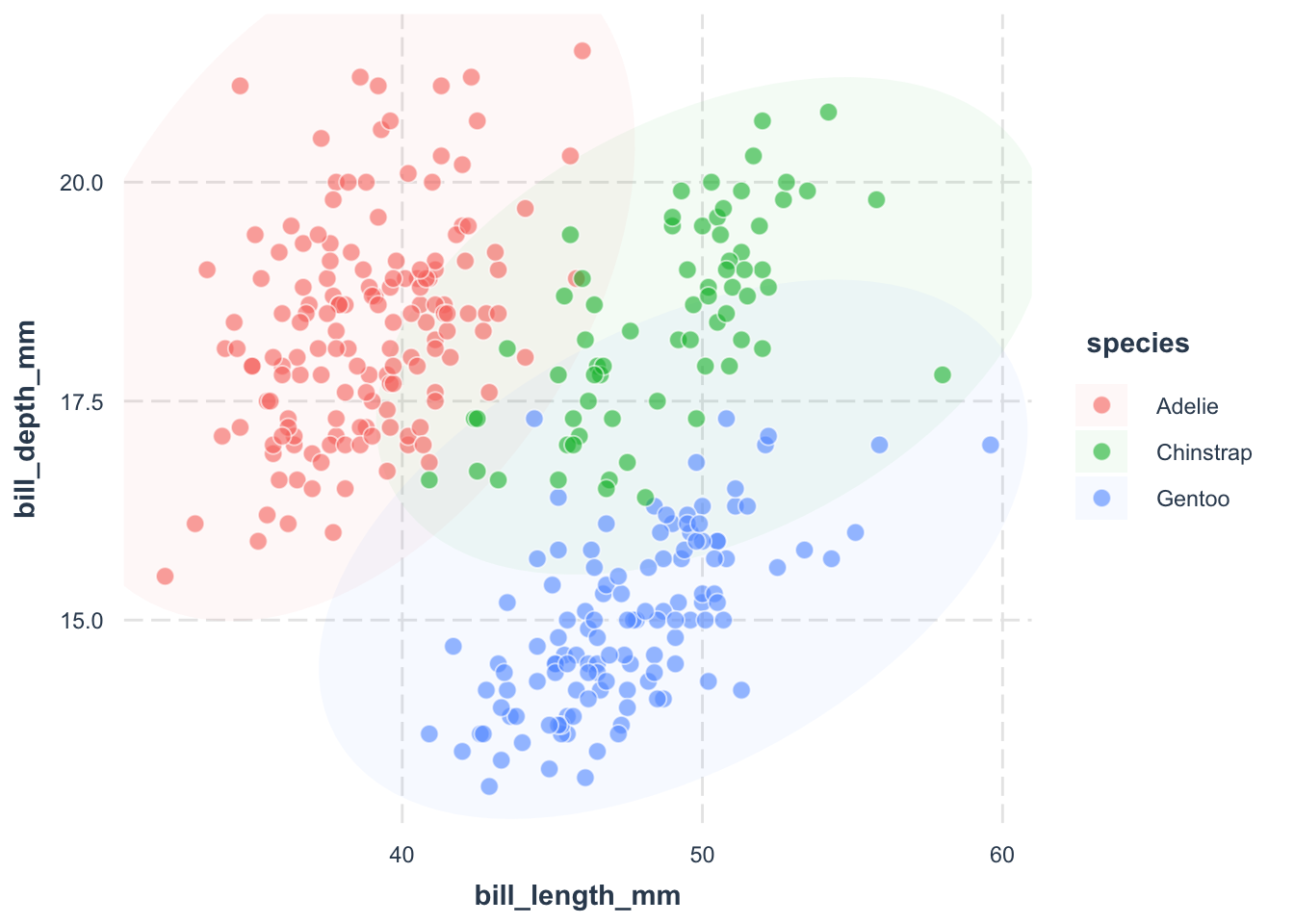
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(
aes(
fill = species,
size = body_mass_g
),
shape = 21,
color = "transparent",
alpha = .3
) +
geom_point(
aes(
size = body_mass_g
),
shape = 21,
color = "white",
fill = "transparent"
) +
jtools::theme_nice()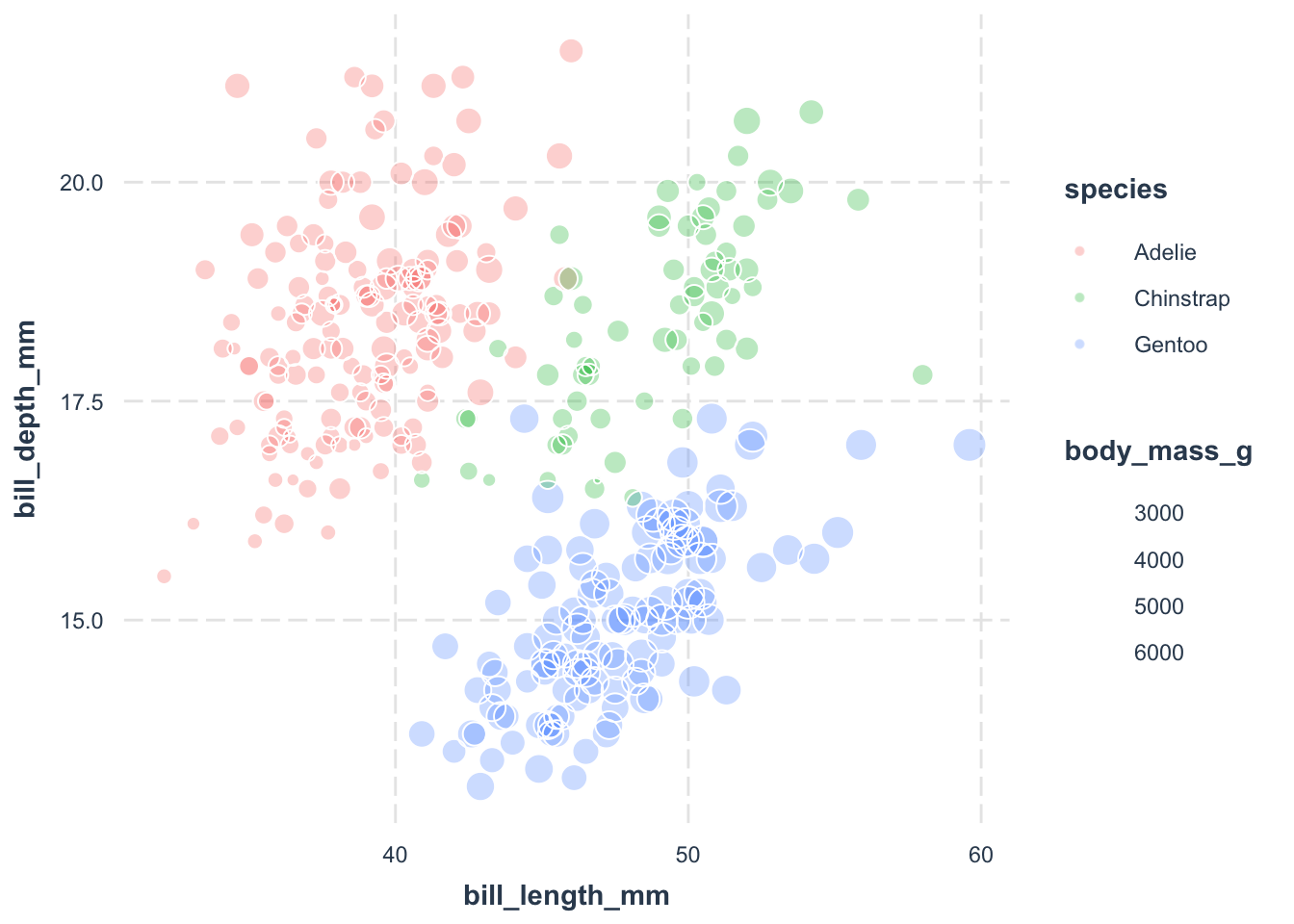
We’ve got points colored by species. However, when people print it out in black and white, they may not be able to distinguish the points. Let’s assign different shapes to each species for better clarity. To be concistent, we use other hollow shapes for points.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
ggforce::geom_mark_ellipse(
aes(fill = species),
alpha = 0.05,
color = 'transparent'
) +
geom_point(
aes(
shape = species,
fill = species
),
color = "white",
size = 3,
alpha = .6
) +
scale_shape_manual(values = c(21, 22, 23)) +
jtools::theme_nice()fill = species argument.
scale_shape_manual(values = c(21, 22, 23)) argument. Nobody can remeber the meanings of these numbers, so just google when you need to.
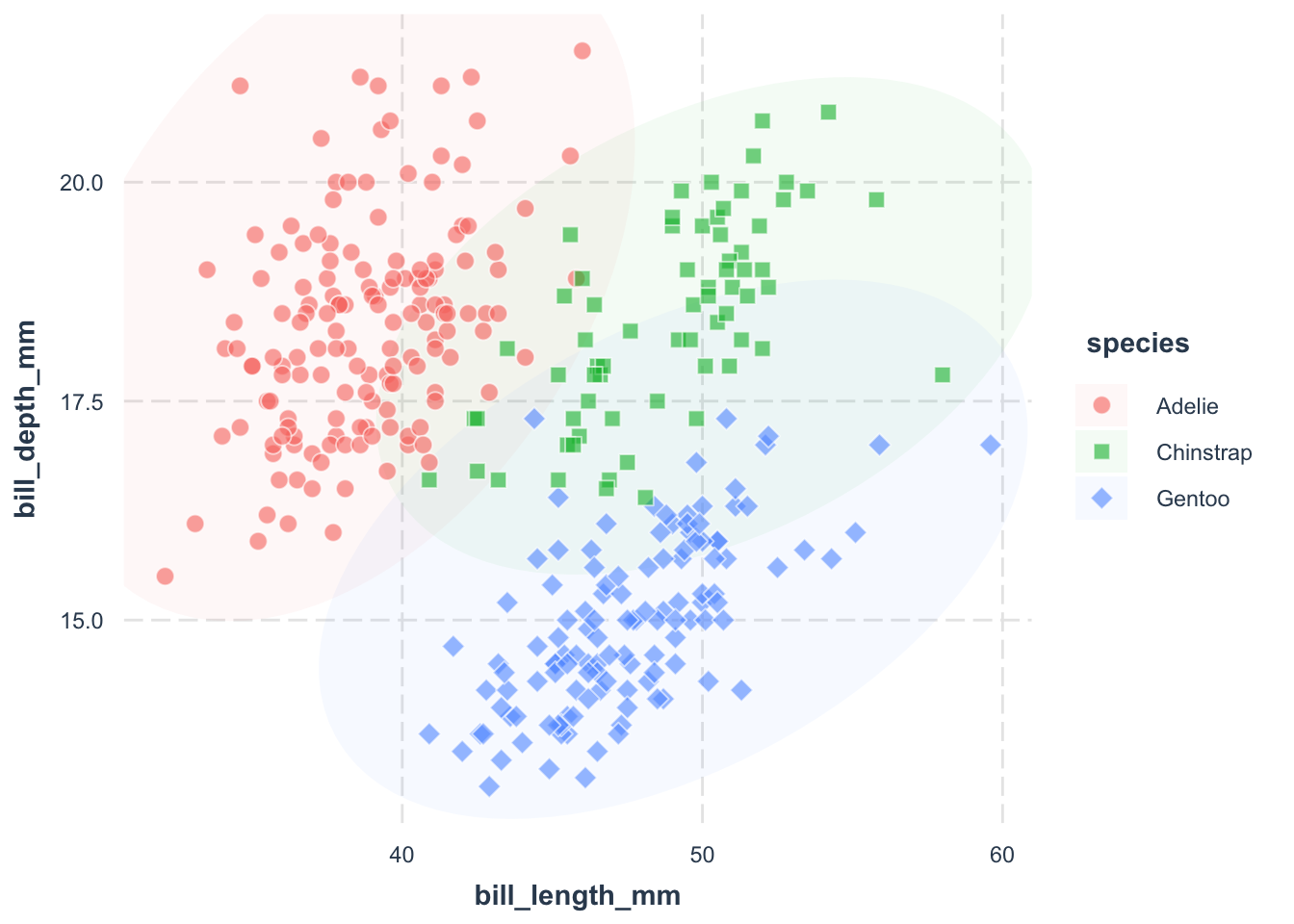
Now, points are distinguishable by both color and shape. Voilà!
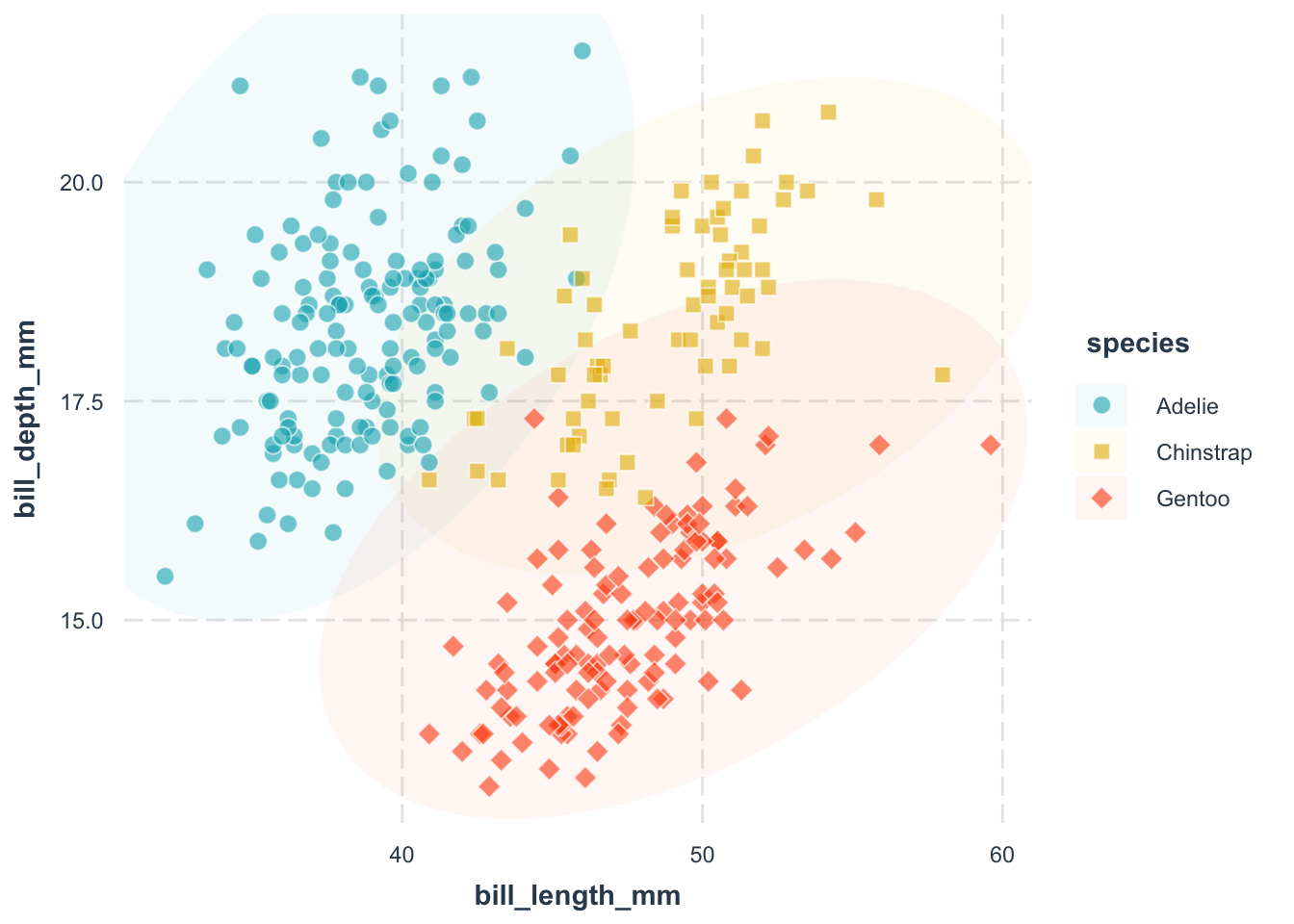
If you are unhappy with the default color palette, you can change it. Here, we use the scale_fill_manual() function to specify the fill color of each species:
library(ggplot2)
library(palmerpenguins)
p <- ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
ggforce::geom_mark_ellipse(
aes(fill = species),
alpha = 0.05,
color = 'transparent'
) +
geom_point(
aes(
shape = species,
fill = species
),
color = "white",
size = 3,
alpha = .6
) +
scale_shape_manual(values = c(21, 22, 23)) +
scale_fill_manual(
values = c(
"Adelie" = "#00AFBB",
"Chinstrap" = "#E7B800",
"Gentoo" = "#FC4E07"
)
) +
jtools::theme_nice()
pscale_fill_manual() function.
Adelie = "#00AFBB" argument.
Chinstrap = "#E7B800" argument.
Gentoo = "#FC4E07" argument.
library(ggplot2)
library(palmerpenguins)
ggplot(
data = penguins,
aes(
x = bill_length_mm,
y = bill_depth_mm
)
) +
geom_point(
aes(fill = species),
color = "white",
shape = 21,
alpha = .6,
size = 3
) +
scale_fill_manual(
values = c("#00AFBB", "#E7B800", "#FC4E07")
) +
jtools::theme_nice()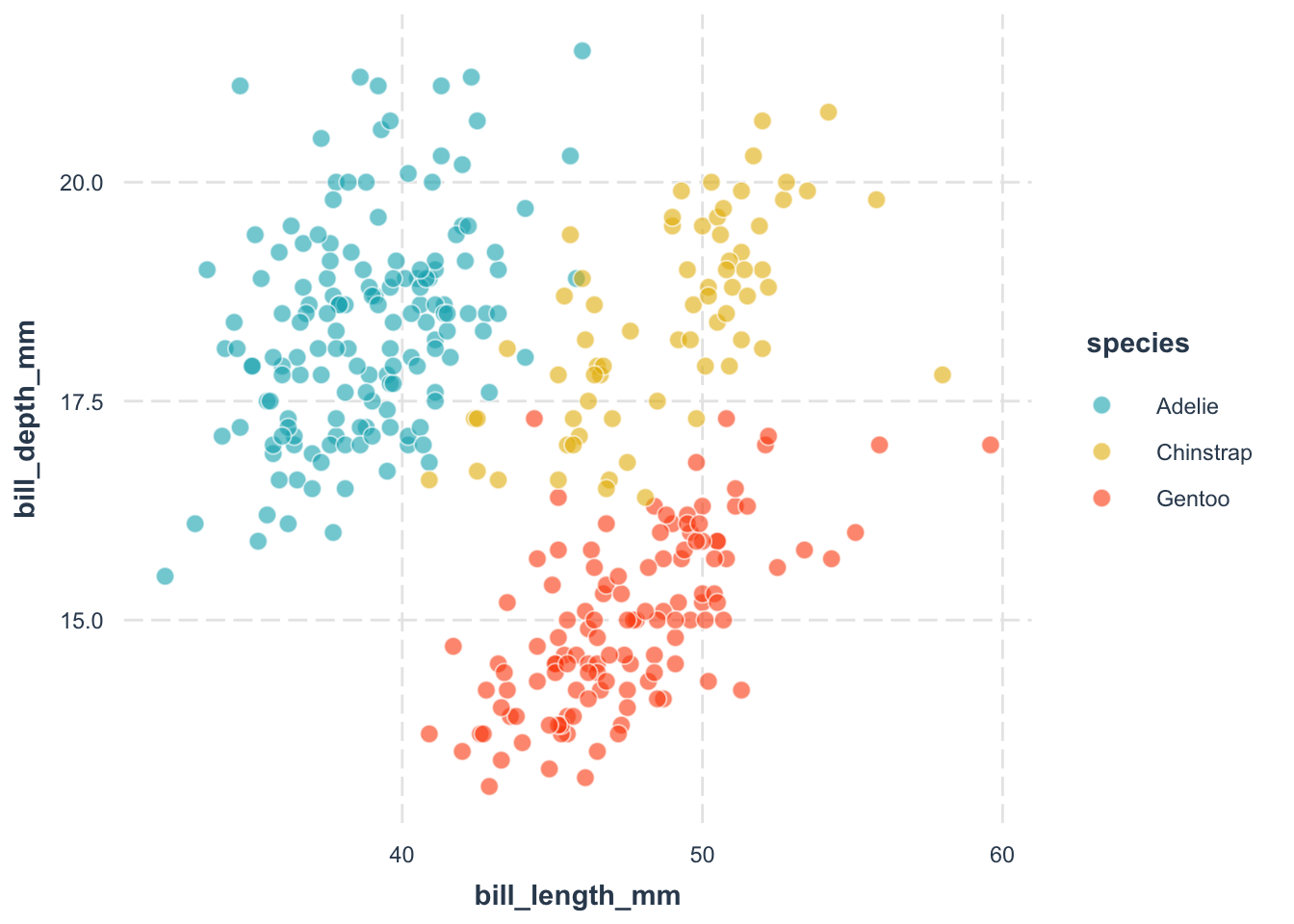
While we have saved a few lines of code, it is not recommended. It is better to use the specific names as keys, so that you can be absolutely sure what color is assigned to which species.
Choose colors wisely. Everyone has their own preferences, but there are some guidelines for best practice. We will get back to this topic later. But for now, I recommend some fun and artsy palettes:
And if you are adventurous, almost all palettes are accessible from the paletteer package (link).
Let us apply what we have learnt so far. Below is a simple scatter plot of the iris dataset. Your task is to customize the plot with:
ggforce::geom_mark_ellipse(), ggforce::geom_mark_hull() and ggforce::geom_mark_rect())library(ggplot2)
ggplot(
data = iris,
aes(
x = Sepal.Width,
y = Petal.Width
)
) +
ggforce::geom_mark_rect(
aes(fill = Species),
alpha = 0.05,
color = 'transparent'
) +
geom_point(
aes(
shape = Species,
fill = Species
),
color = "white",
size = 3,
alpha = .6
) +
scale_shape_manual(values = c(21, 22, 23)) +
scale_fill_manual(
values = MetBrewer::met.brewer(name="Demuth", n=3, type="discrete")
) +
theme_minimal()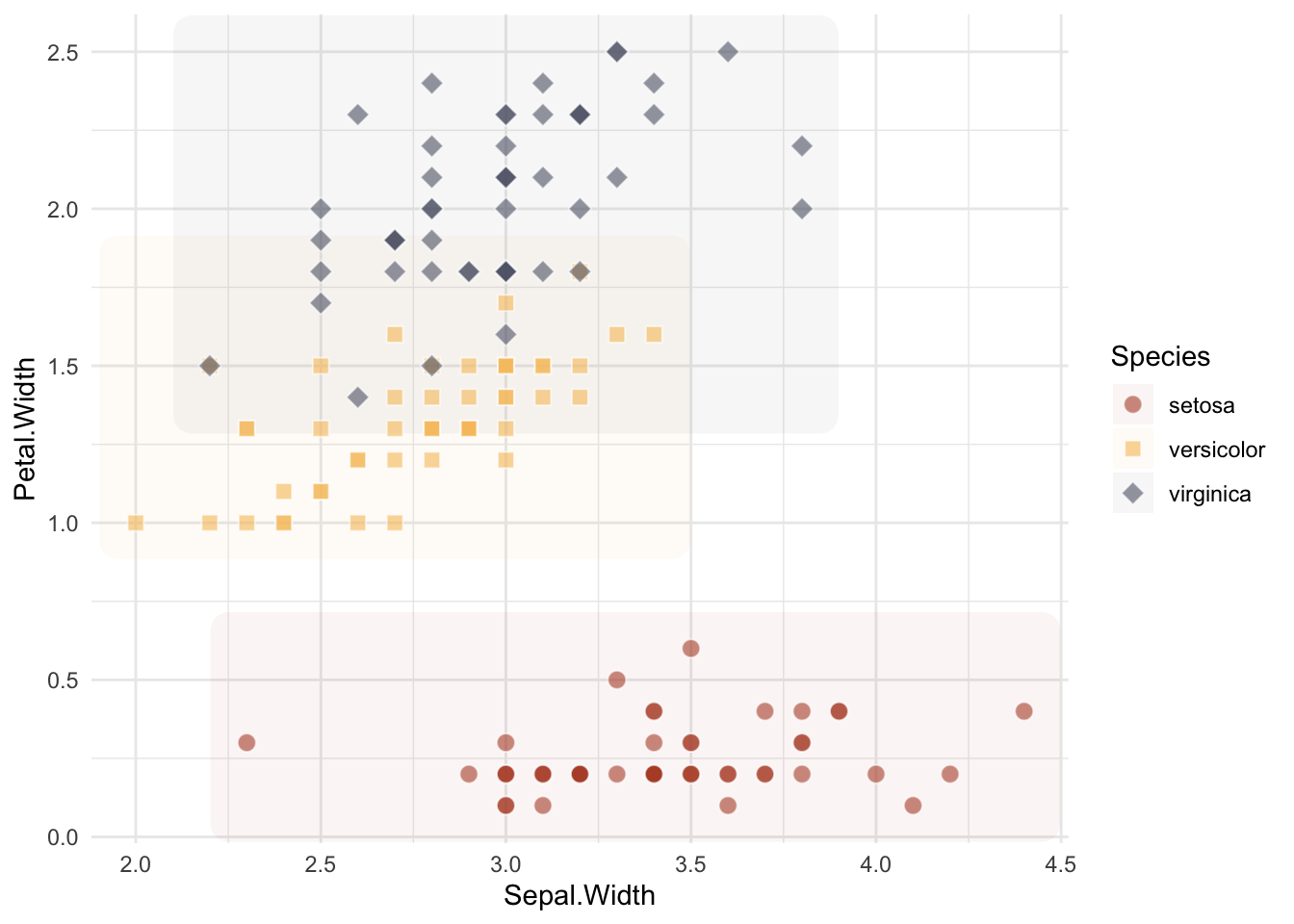
The rule of thumb is to make your plot as self-contained as possible. Ideally, the reader should be able to understand the plot without referring to the text. There are many steps to achieve this, but we will focus on the most important ones for now.
Clarity starts with clear labels. Use the labs() function to name your axes.
p +
labs(
x = "Bill Length (mm)",
y = "Bill Depth (mm)"
)x = "label text" argument.
y = "label text" argument.
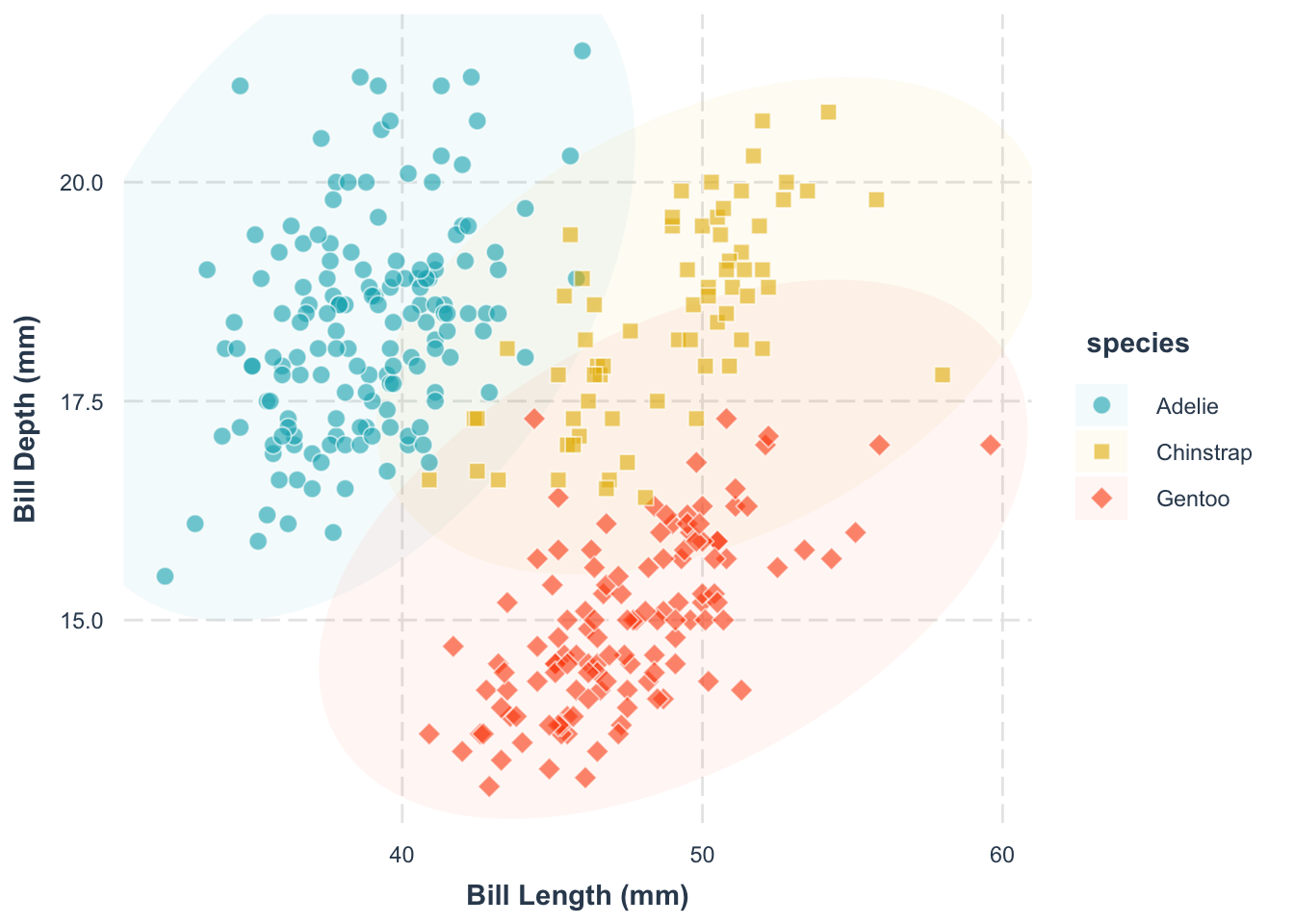
We will get back to labelling later (with the library ggtext and ggrepel), but for now, let’s keep it simple.
Notice how we add the layer directly to the plot p from the previous figure. This is a common (and very powerful) trick in ggplot2.
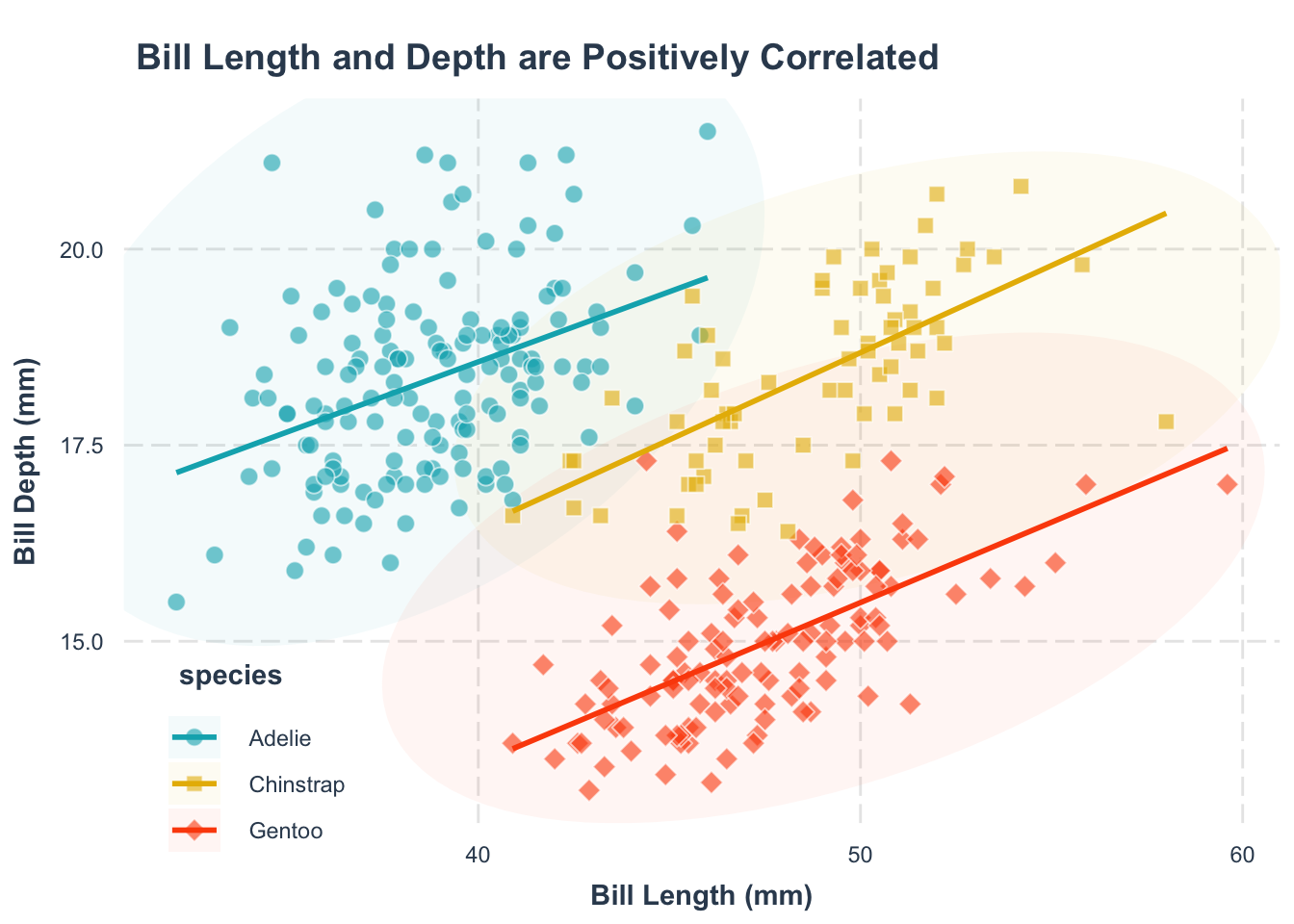
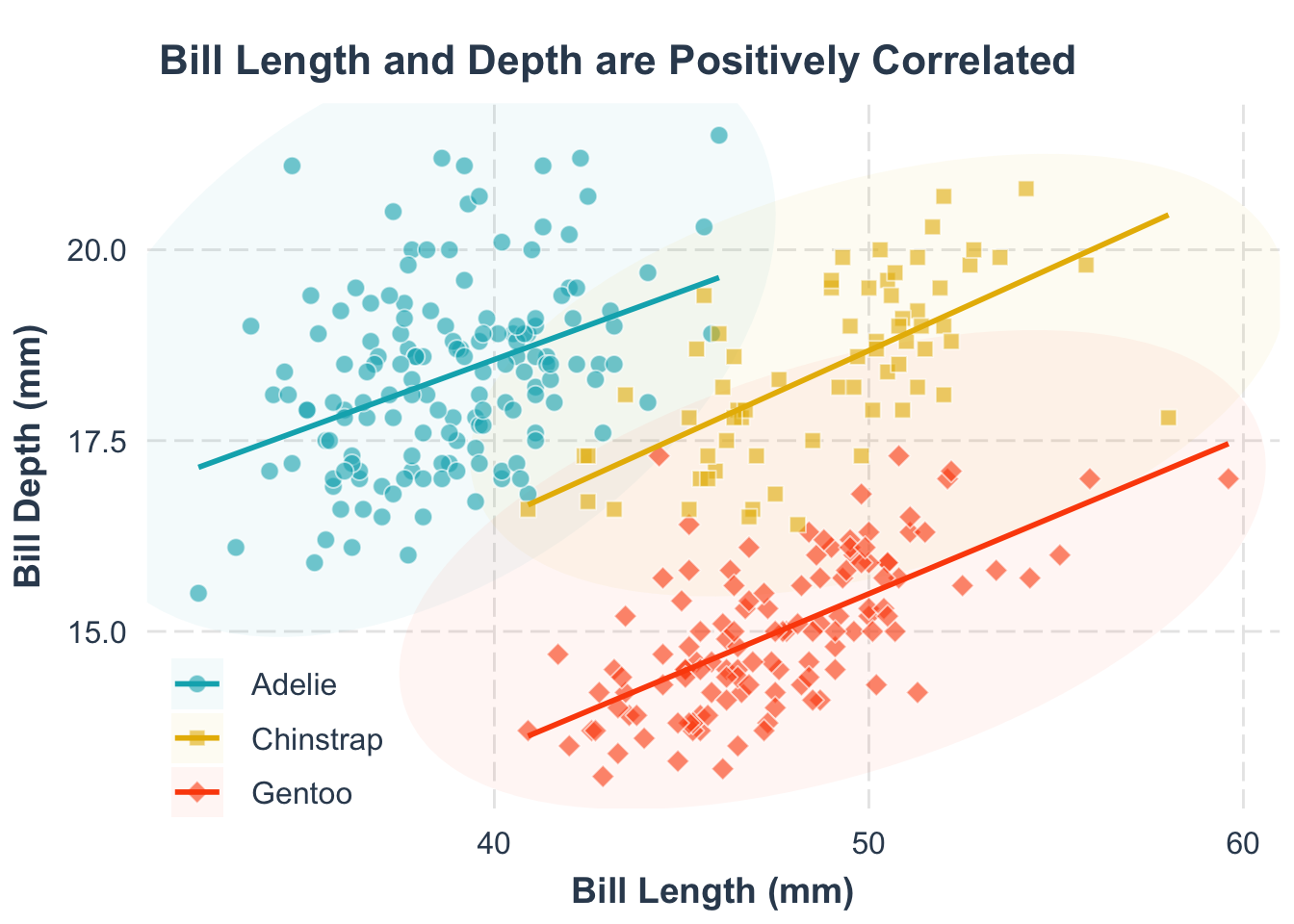
Second, what’s this plot telling us? A title can make that clear, also using labs():
p +
labs(
x = "Bill Length (mm)",
y = "Bill Depth (mm)",
title = "Bill Length and Depth of Penguins are Positively Correlated"
)title = "title text" argument.
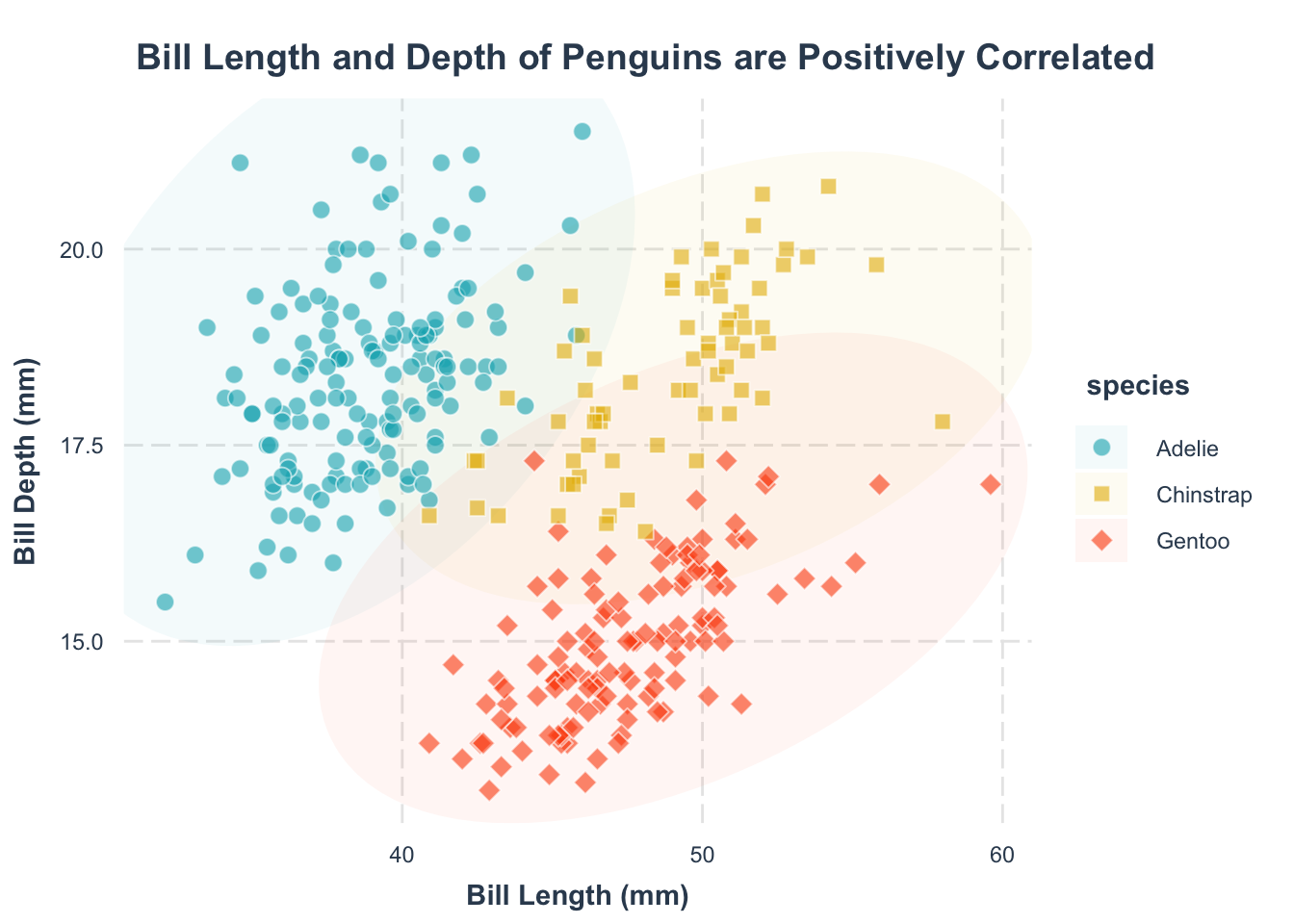
Now, your plot clearly communicates its main message.
Help your readers spot trends by adding a trend line. Remember, the goal is to make the plot as easy to understand as possible.
p1 <- p +
labs(
x = "Bill Length (mm)",
y = "Bill Depth (mm)",
title = "Bill Length and Depth are Positively Correlated"
) +
geom_smooth(
aes(group = species, color = species),
method = "lm", se = FALSE
) +
scale_color_manual(
values = c(
"Adelie" = "#00AFBB",
"Chinstrap" = "#E7B800",
"Gentoo" = "#FC4E07"
)
)
p1geom_smooth() function. The group = species argument specifies that we want to fit a separate trend line for each species.
geom_smooth() function. The method = "lm" argument specifies that we want to fit a linear model to the data. The se = FALSE argument specifies that we do not want to display the standard error around the trend line.
scale_color_manual() function.
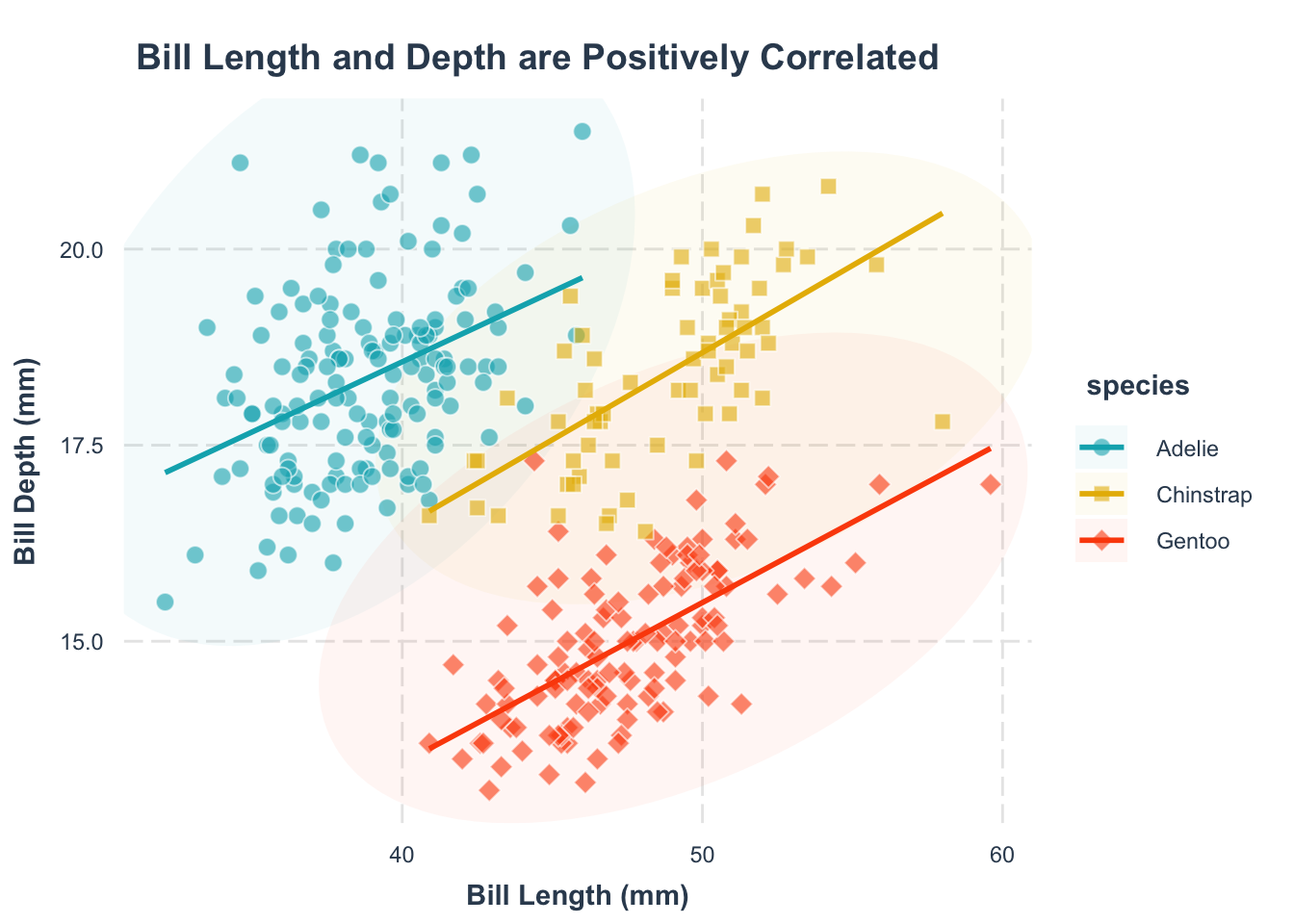
Note: color and fill are different aesthetics in ggplot2, so you need to set them separately. The grammar for setting them is the same.
Let us apply what we have learnt so far. Below is a simple scatter plot of the iris dataset. Your task is to customize the plot with customized point shape for each species and use a non-default color.
library(ggplot2)
library(MetBrewer)
p_exe <- ggplot(
data = iris,
aes(
x = Sepal.Width,
y = Petal.Width
)
) +
geom_point(
aes(
shape = Species,
fill = Species
),
color = "white",
size = 3,
alpha = .6
) +
scale_shape_manual(values = c(21, 22, 23)) +
scale_fill_manual(
values = MetBrewer::met.brewer(name="Demuth", n=3, type="discrete")
) +
theme_minimal()
p_exe +
labs(
x = "Petal width (cm)",
y = "Sepal width (cm)",
title = "Association between Sepal and Petal Width"
) +
geom_smooth(
aes(group = Species, color = Species),
method = "lm", se = FALSE
) +
scale_color_manual(
values = MetBrewer::met.brewer(name="Demuth", n=3, type="discrete")
)`geom_smooth()` using formula = 'y ~ x'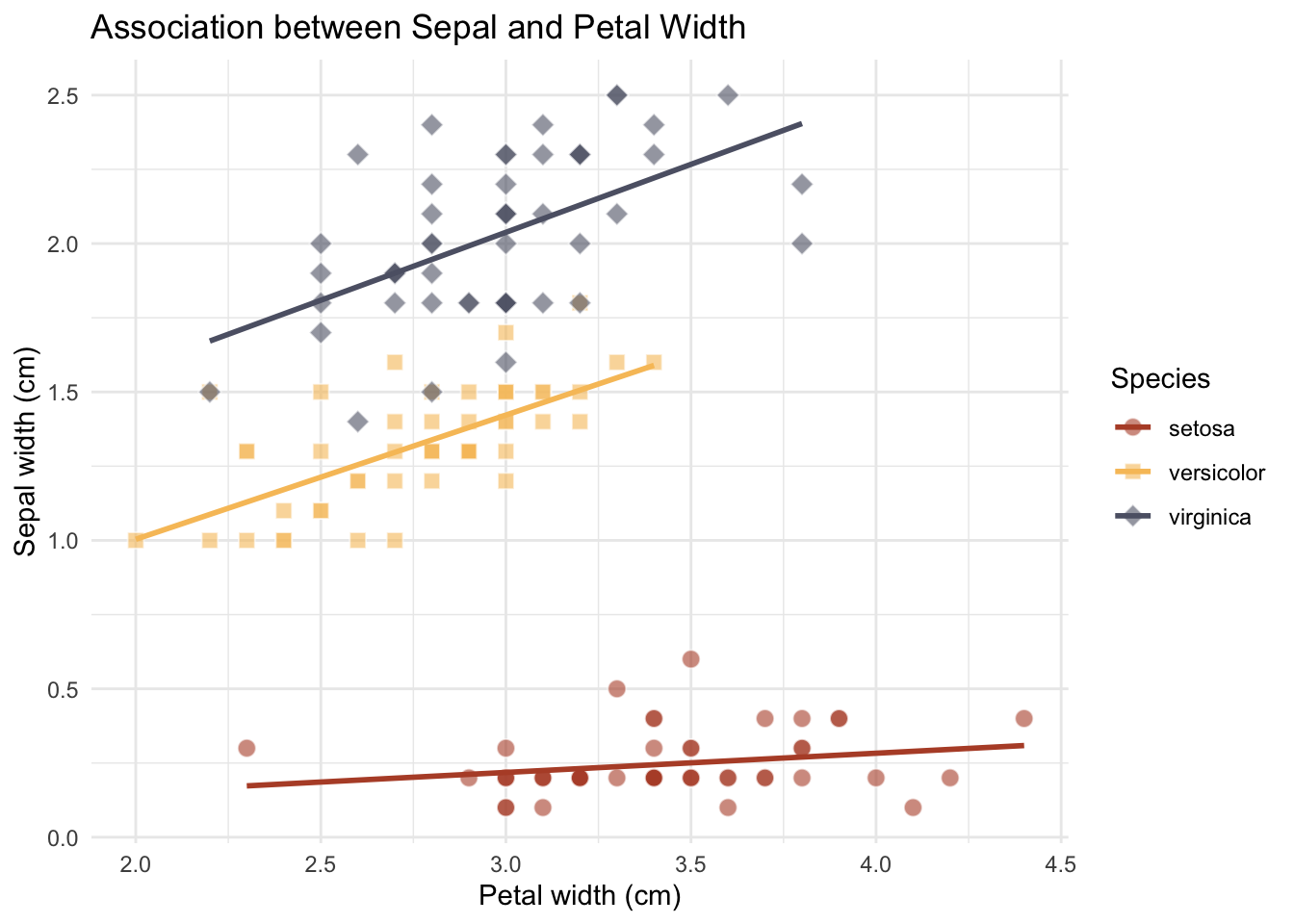
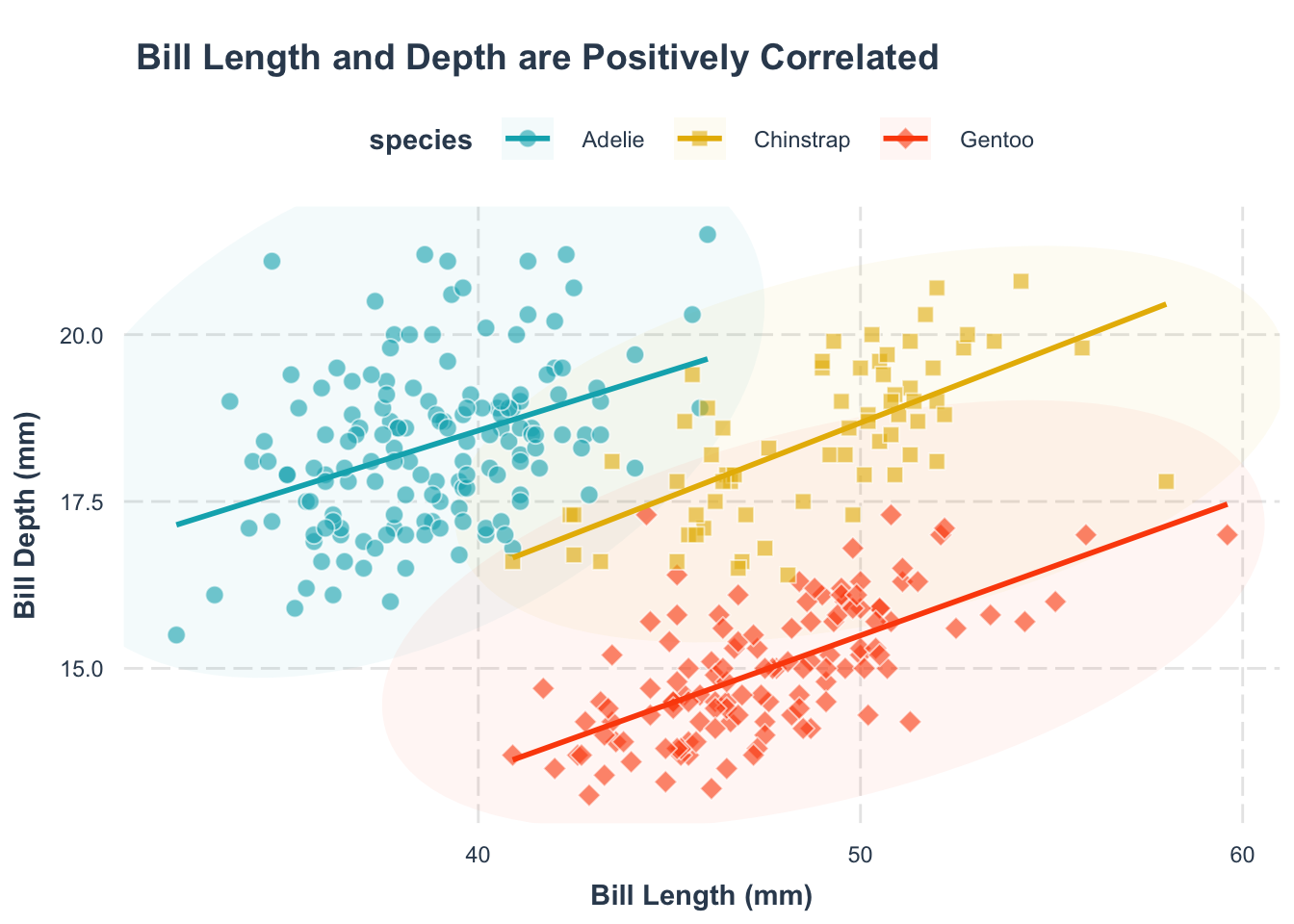
By default, the legend hangs out on the right. Often, placing it on top (or bottom) or inside the figure makes for a cleaner look. Let’s see how.
p1 +
theme(
legend.position = c(0.12, 0.1)
)legend.position = c(*, *) argument.
The first number is the x-coordinate, and the second is the y-coordinate, both ranging from 0 (left/bottom) to 1 (right/top).
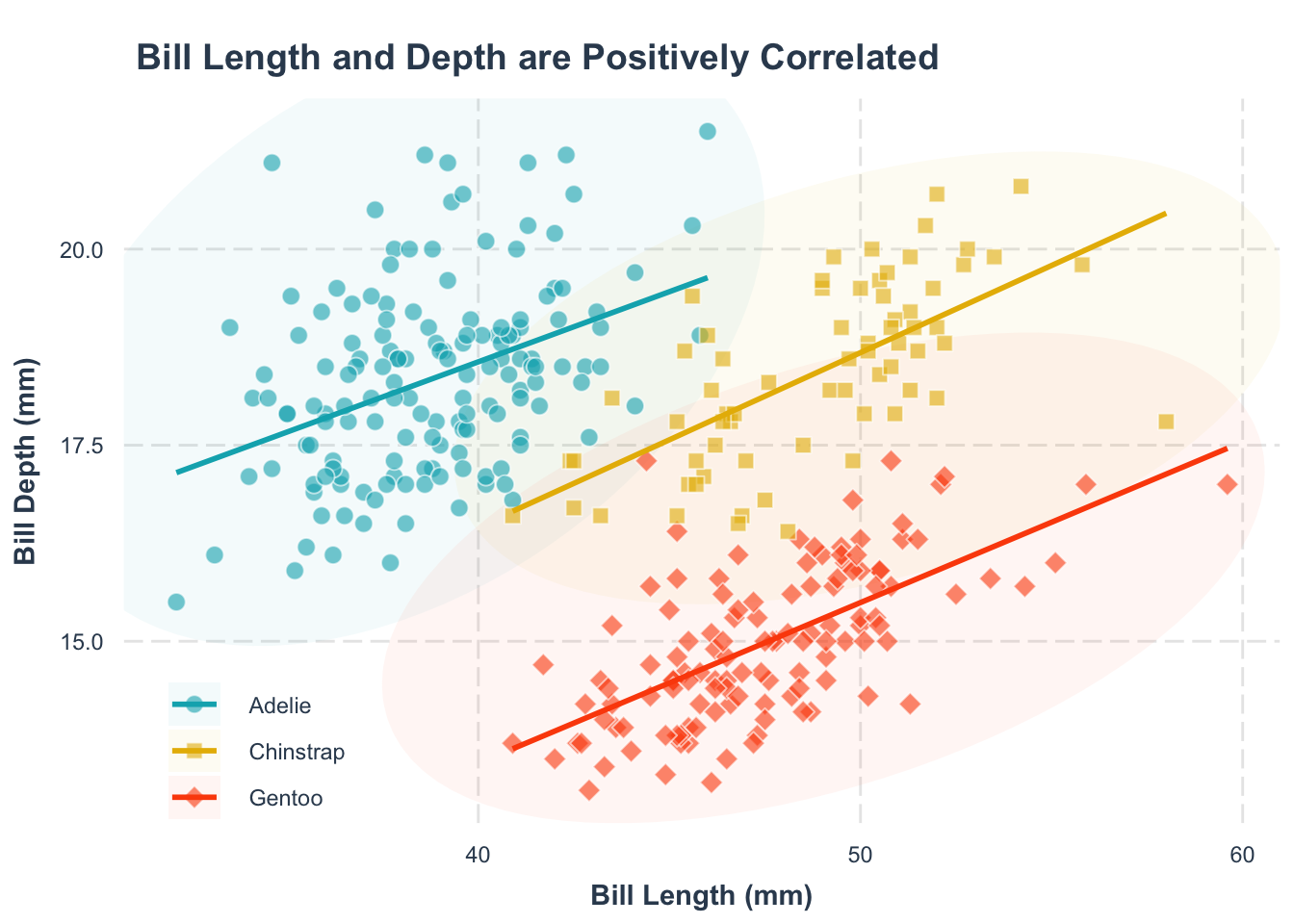
p1 +
theme(
legend.position = "top"
# legend.position = "bottom"
)legend.position = "top" argument.
legend.position = "bottom" argument.
Another common blunder: not paying attention to the legend title.
Sometimes, for example with expert audiences, the legend title is redundant. Remove it with theme()
p2 <- p1 +
theme(
legend.position = c(0.12, 0.1),
legend.title = element_blank()
)
p2legend.title = element_blank() argument.
For a more informative, self-contained figure, you might need to rewrite the legend title. Since ggplot2 can merge multiple legends (color, shape, linetype), ensure consistency by renaming all relevant legends.
p1 +
theme(
legend.position = "top"
) +
labs(
color = "Penguin species",
shape = "Penguin species",
fill = "Penguin species", # <3>,
linetype = "Penguin species"
)labs(color = "label text") argument.
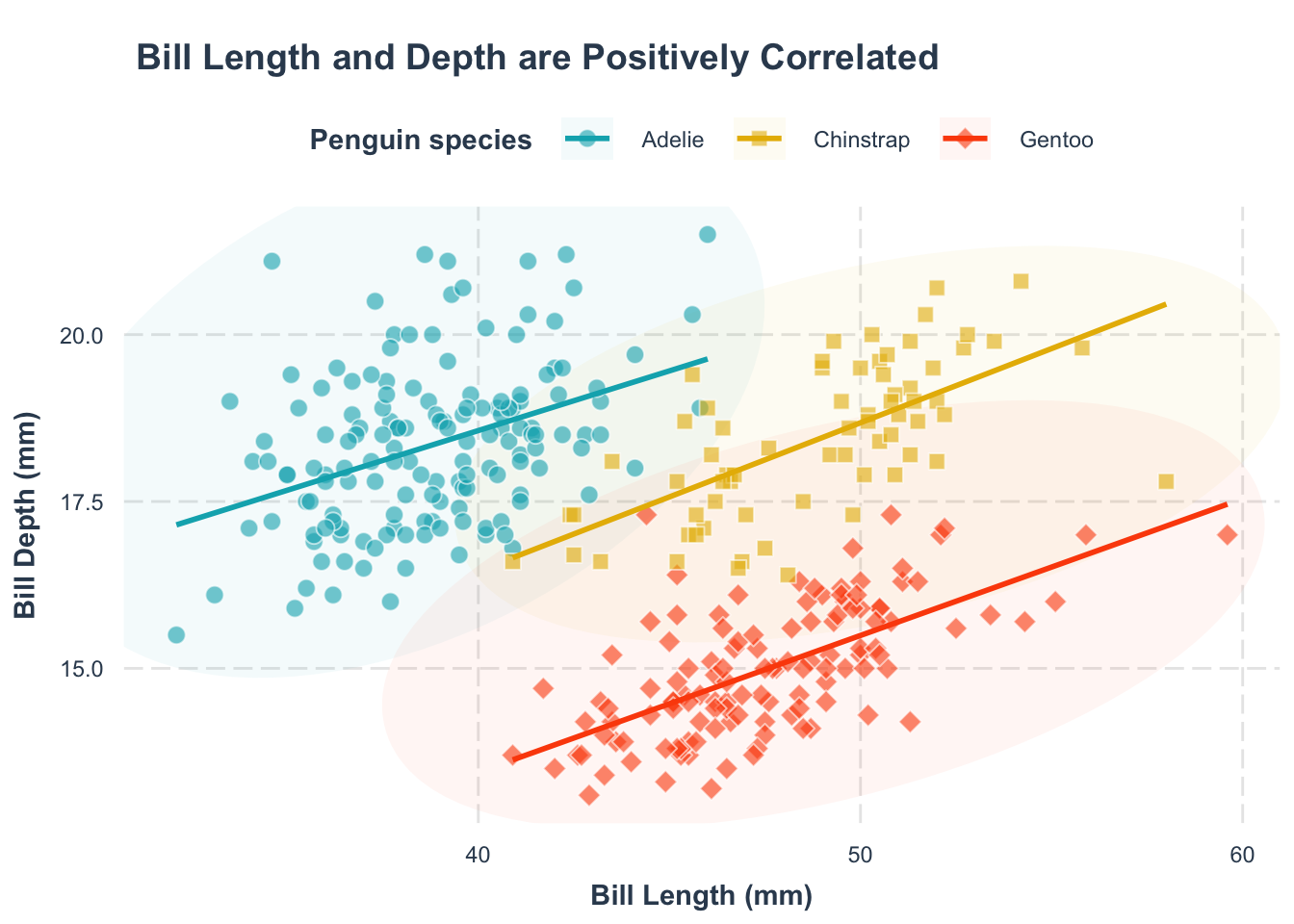
Another approach is to design the figure so that the legend is not necessary. They are usually more complicated and labor-heavy, but definitely worth it. Below we show two approaches:
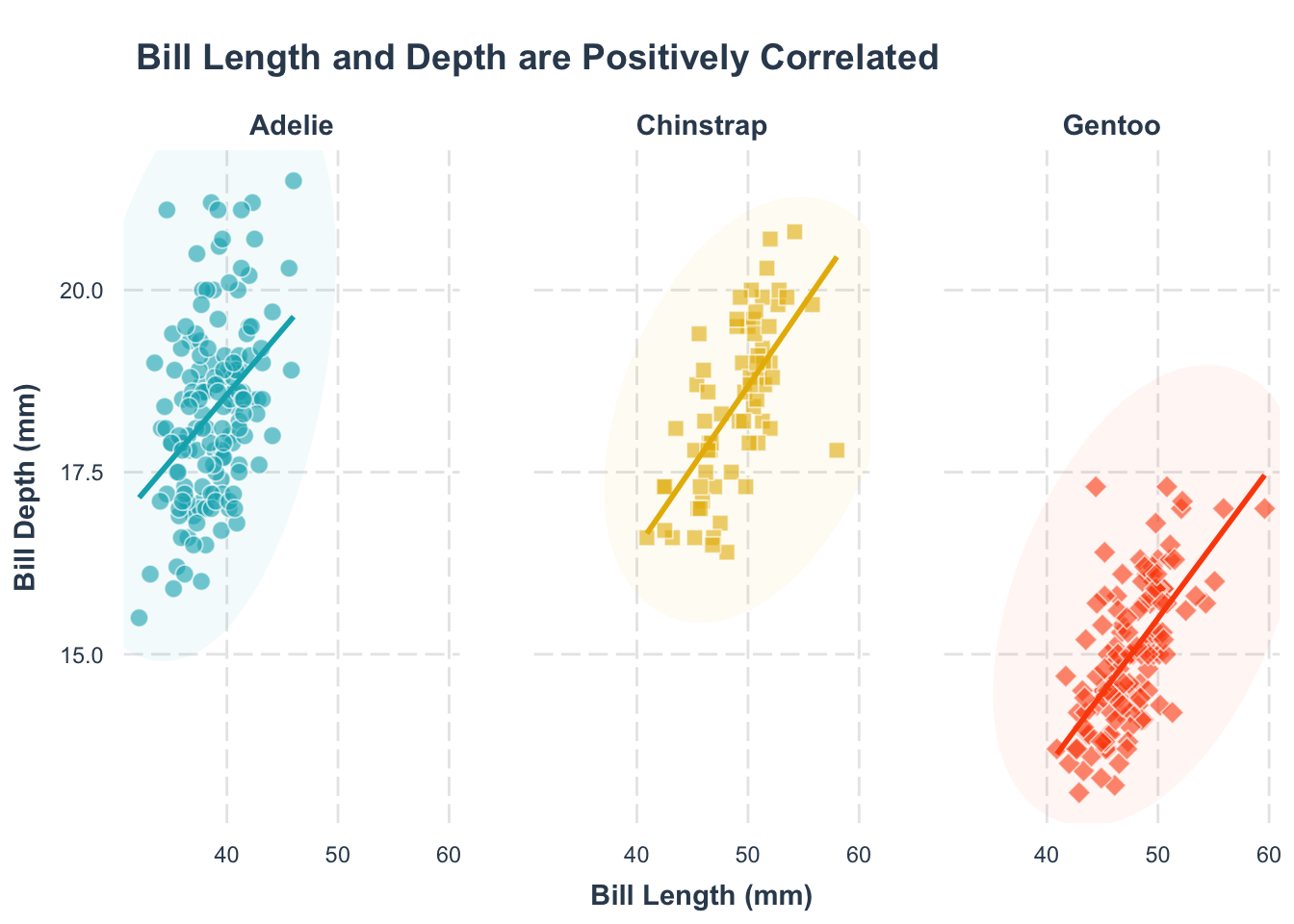
p1 +
facet_wrap(~species) +
theme(
legend.position = "none"
)
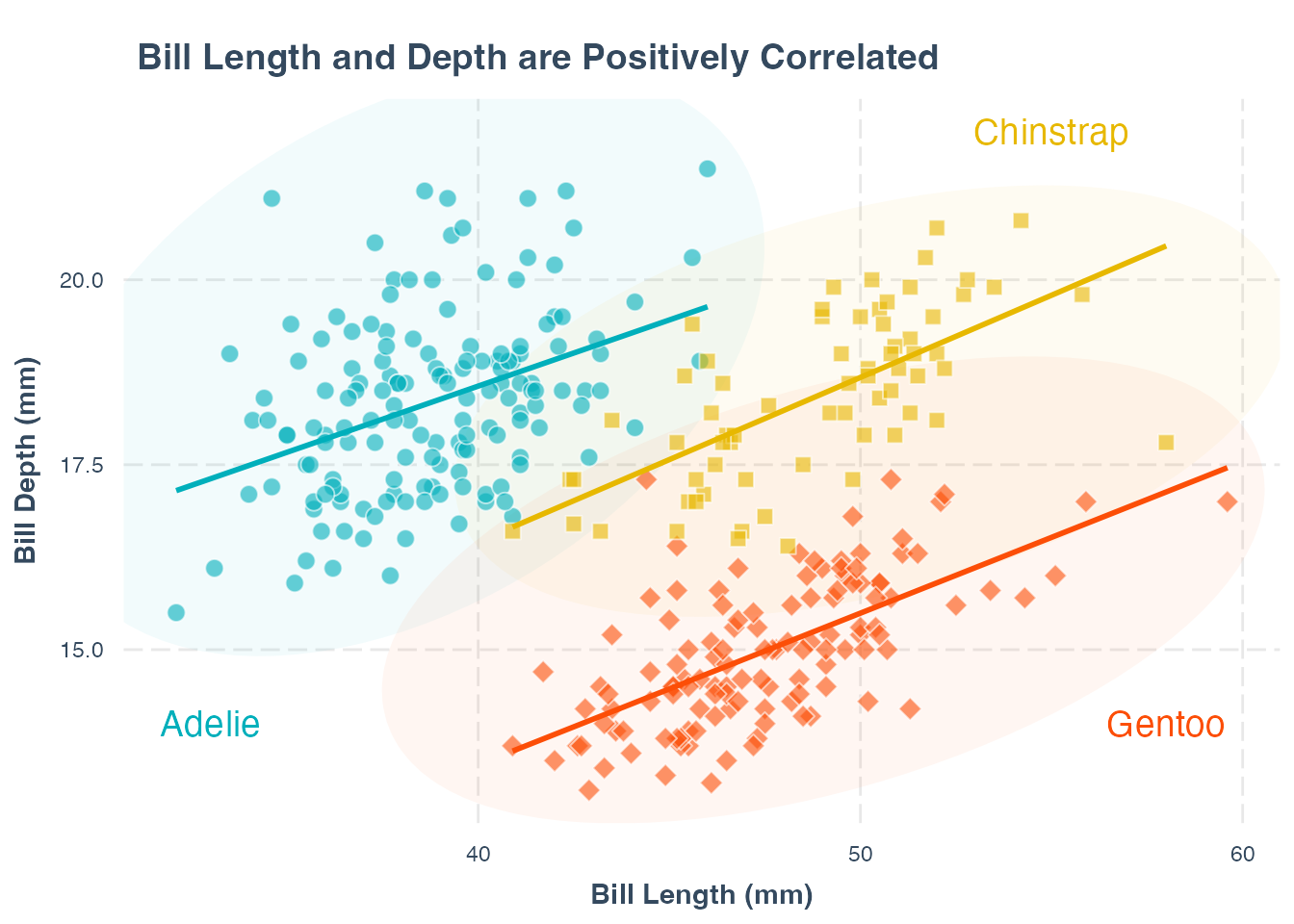
p1 +
theme(legend.position = "none") +
annotate("text",
x = 33, y = 14,
label = "Adelie",
color = "#00AFBB", size = 5
) +
annotate("text",
x = 55, y = 22,
label = "Chinstrap", color = "#E7B800", size = 5
) +
annotate("text",
x = 58, y = 14,
label = "Gentoo", color = "#FC4E07", size = 5
)annotate() function.
x = , y = arguments.
label = "label text" argument.
color = argument. We increase the size of the text annotations using the size = argument.
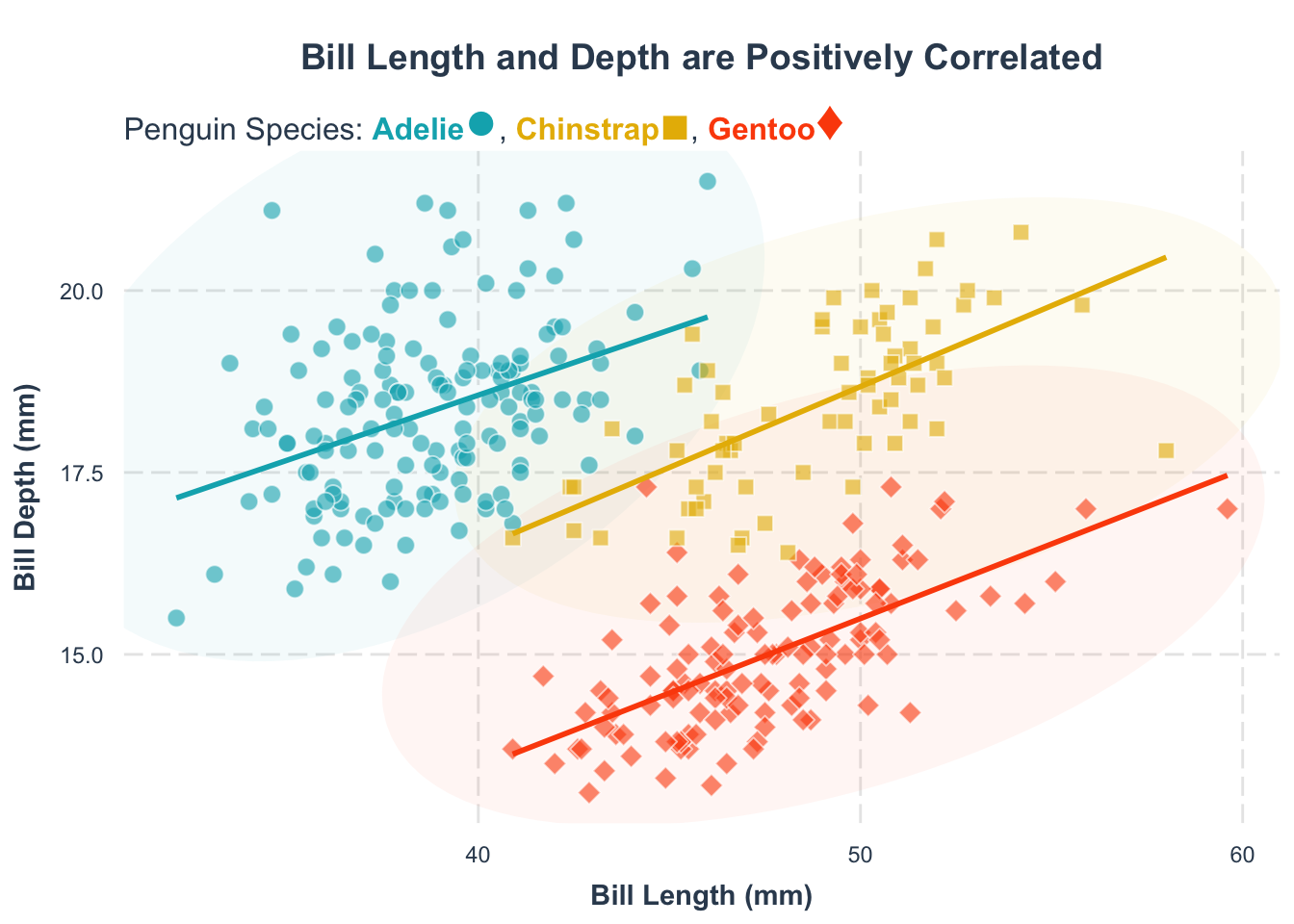
We can use subtitle as an effective way to label the groups. We can use the ggtext package to add the subtitle. If you have not installed the package yet, just run install.packages("ggtext").
library(ggtext)
p1 +
theme(
legend.position = "none"
) +
labs(
subtitle = "Penguin Species:
<span style = 'color:#00AFBB;'>**Adelie**</span><span style = 'color:#00AFBB;font-size:22pt'>\u25CF</span>,
<span style = 'color:#E7B800;'>**Chinstrap**</span><span style = 'color:#E7B800;font-size:20pt'>\u25A0</span>,
<span style = 'color:#FC4E07;'>**Gentoo**</span><span style = 'color:#FC4E07;font-size:22pt'>\u2666</span>"
) +
theme(
plot.title = element_text(hjust = 0.5),
plot.subtitle = element_textbox_simple(halign = 0, size = 12)
)
There is also another package marquee that makes this process easier. But it is still under active development. Check the documentation if you are intrested.
Whenever I read papers or listen to talks, I often see that the font size is too small. The following is a quote from the book Fundamentals of Data Visualization by Claus Wilke:
If you take away only one single lesson from this book, make it this one: Pay attention to your axis labels, axis tick labels, and other assorted plot annotations. Chances are they are too small. In my experience, nearly all plot libraries and graphing softwares have poor defaults. If you use the default values, you’re almost certainly making a poor choice.
We can increase the font size of the axis labels using the theme() function.
p3 <- p2 +
theme(
axis.text = element_text(size = 12),
axis.title = element_text(size = 14),
plot.title = element_text(size = 16),
legend.text = element_text(size = 12)
)
p3theme() function.
axis.text = element_text(size = ) argument.
axis.title = element_text(size = ) argument.
title = element_text(size = ) argument.
legend.text = element_text(size = ) argument.
Selecting the appropriate font size in ggplot2 is somewhat of an art. A common mistake is using a font size that looks great in the RStudio preview but doesn’t scale appropriately when exporting the plot to different sizes. While the plot elements typically adjust to the export dimensions, fixed font sizes do not, leading to readability issues and challenges with reproducibility. To address this,
p2 +
patchwork::plot_layout(widths = 50, heights = 50) +
theme(...)Alright, brace yourself. The full code for our pièce de résistance—our final figure—is right below. Run it with a single click and marvel as the figure above magically appears:
Yes, the code might look like it’s speaking in tongues, but fear not! It’s all about the logic. Let’s break down the wizardry step by step:
ggplot() function. This is your canvas. Specify your data and map out the x and y axes.geom_*). What to add are motivated by what we want to show. Since we’re exploring the relationship between bill length and bill depth, we need:
geom_point): Each penguin observation.geom_smooth): To highlight that positive relationship.ggforce::geom_mark_ellipse): To neatly separate different species.scale_* function. In this plot, the customized aes includes shape, color, and fill.labs(): Clear labels make your plot understandable at a glance.theme() function. We can change the font size, the position of the legend, etc.The logic behind every ggplot2 figure follows these structured steps. It may seem intimidating at first, but I promise you will get used to it and even fall in love with it:
Finally, we need to export the plot such as pdf or png, so we can use it elsewhere (like submit to a journal). However, a minimalist approach like ggsave("plot_name.pdf") lacks the precision needed for publication-quality figures. Below, we show the best practices for exporting your plot:
PDFs are the gold standard for vector graphics in academic publications due to their scalability and clarity. To save your plot as a PDF, consider the following comprehensive method:
ggsave(
filename = "plot.pdf",
plot = p3,
width = 8,
height = 6,
units = "in",
device = cairo_pdf
)Why do we bother to specify plot dimensions? For one thing, RStudio window sizes can vary from one computer to another, which may cause your figures to appear distorted or improperly sized if dimensions aren’t fixed. Additionally, font sizes in plots are fixed and do not automatically scale with plot dimensions, leading to text that may be too large or too small relative to the plot size. I’ve had nightmares to reproduce well-formatted figures because I forgot to set these dimensions. To avoid such headaches, it’s best practice to always specify the width and height when exporting your plots.
While PDFs are ideal for vector graphics, many journals only accept non-vector format like PNGs. To achieve optimal quality and avoid common pitfalls related to fonts and rendering, the ragg package is highly recommended. If you haven’t installed it yet, simply execute install.packages(“ragg”).
ggsave(
filename = "plot.png",
plot = p3,
width = 8,
height = 6,
units = "in",
device = ragg::agg_png
)While ggplot2 is powerful, it is not a design software and it can be paiful to do everything in R. To add extra touch, you can save it as a vector graphic (e.g., pdf) and edit it in a design software like Adobe Illustrator or Inkscape.
However, learning a new software can be time-consuming. Luckily, you get to do many of these tasks in PowerPoint. The trick is to save the plot as svg format. This is a vector graphic format that can be easily imported into editable format in PowerPoint. Another practicle trick is to remove the white background of the plot. Below is the code to do this:
p_svg <- p3 +
theme(
panel.background = element_rect(fill = "transparent", colour = NA_character_),
plot.background = element_rect(fill = "transparent", colour = NA_character_),
legend.background = element_rect(fill = "transparent"),
legend.box.background = element_rect(fill = "transparent"),
legend.key = element_rect(fill = "transparent")
)
ggsave(
filename = "plot.svg",
plot = p_svg,
width = 8,
height = 6,
units = "in"
)With the exported svg in Keynote, you can easily add annotations, arrows, and other design elements to make your plot more informative and appealing. As an example, below adds the artistic plot of penguins to better illustate the two axis:
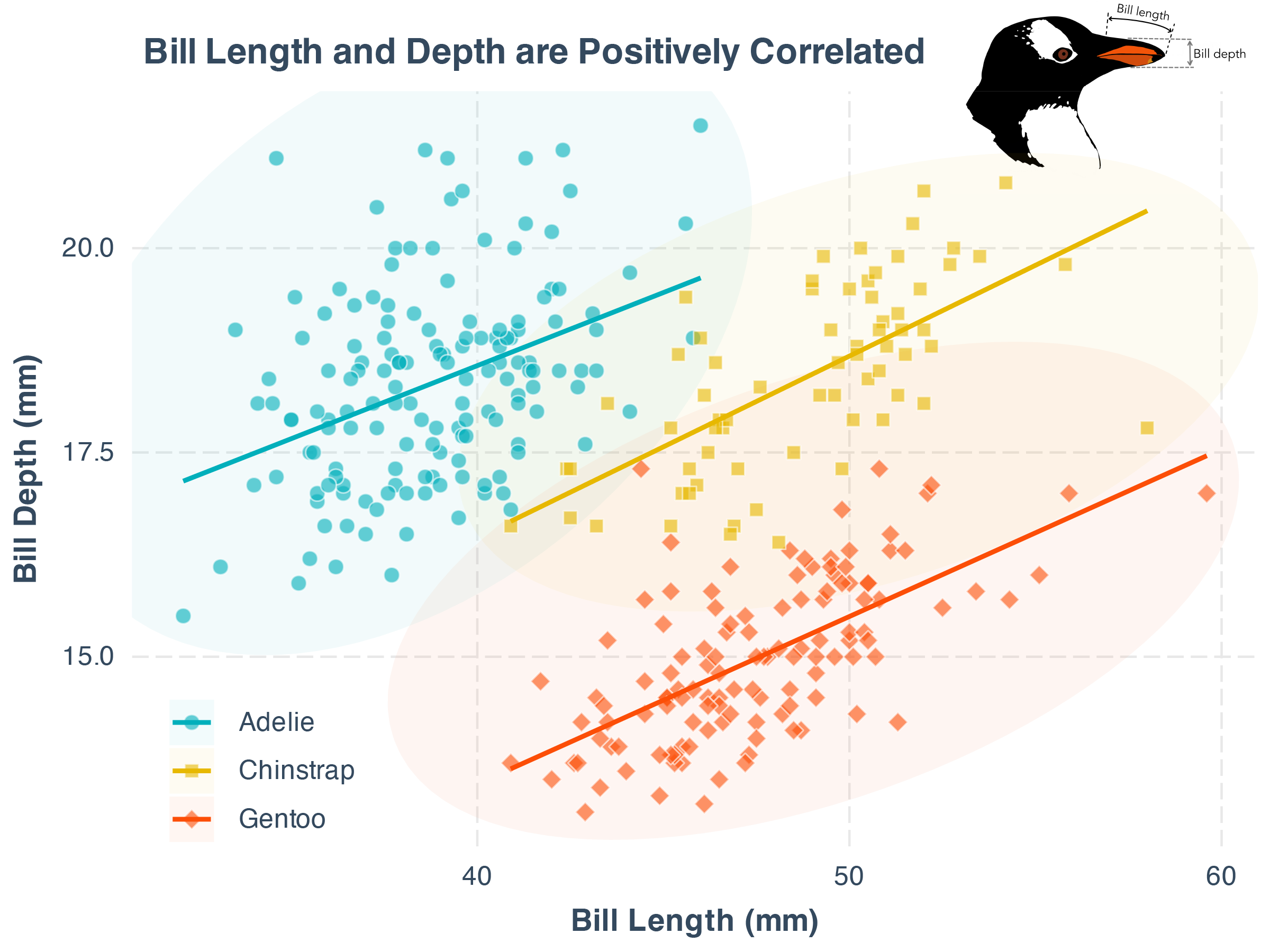
A well-drawn illustration can make your plot more engaging. It is not easy, as this is not a standardized process. That said, here are some solid options for finding awesome illustrations:
Another great side benefit of editing this in Powerpoint is that that you can esaily animate your figures when you present in conferences or group meetings. We will demonstratre in class how to do this.