numbers <- c(1, 2, 3, 4, 5)
numbers- 1
- Create a vector of numbers from 1 to 5.
- 2
-
Display the
numbersvector.
[1] 1 2 3 4 5vector and tibble in RA data structure is an orderly, efficient way to store and retrieve data. While it may feel a bit abstract if you’re new to coding, think of these structures as the “nouns” in your programming language (with data wrangling serving as the “verbs”).
R has provided many data structures. Fortunately, nine times out of ten, you’ll only need to worry about two: vector and tibble.
A vector is a one-dimensional array—like a row of seats in a movie theater that only allows one type of audience member, be they numeric, character, or logical. We create them with the c() function. For example:
numbers <- c(1, 2, 3, 4, 5)
numbersnumbers vector.
[1] 1 2 3 4 5c() stand for
c() means “combine.” Yes, it’s a slightly cryptic name for what you’ll soon be typing all the time, but you’ll get used to it.
Vectors form the backbone of R scripts. They’re how you can store the number of penguins wandering around Antarctica, keep track of class attendance, or tally cars on the highway. A key detail: everything in a vector must share the same type. Some common types are:
numeric (like 3.14 or 42)character (like “Hello” or “World”)logical (TRUE or FALSE)factor (categorical data)Below, we make a few different vectors to illustrate these types:
numbers <- c(1.1, 2.2, 3.3, 4.4, 5.5)
characters <- c("a", "b", "c")
logicals <- c(TRUE, FALSE, TRUE)
factors <- factor(c("a", "b", "c"))R automatically assigns the data type for each vector, which can save time but also lead to (unwanted) surprises. For instance, sometimes 1 appears as a character rather than a number—something that can introduce annoying bugs. Avoid headaches by checking data types with class(). For example:
class(numbers) [1] "numeric"A horror story (not in R but in Excel) is that the auto-convert feature messes up gene names like SEPT4 to 4-Sept (datatype from character to datetime). This has affected a ton of genetic papers. So, always be aware of the data types!
We can also extract data from a vector using the square brackets []. For example:
numbers[1]
characters[2]numbers vector.
characters vector.
[1] 1.1
[1] "b"Note that the index starts from 1, not 0. This is a common source of confusion for those coming from languages like Python or C. I personally prefer the 1-based index, as this is more intuitive.
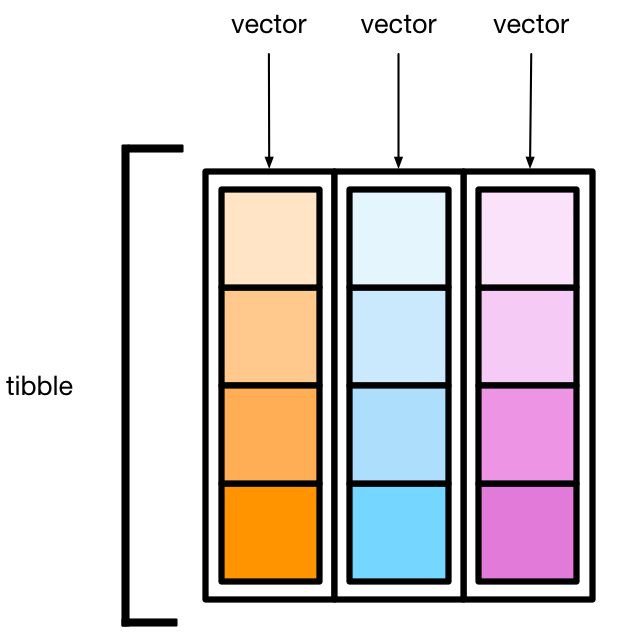
A tibble (brought to you by the tidyverse) is essentially a set of vectors bound together in columns—like a multi-row, multi-column theater, where each column houses a single data type.
As a simple xample,
library(tidyverse)
tibble(
x = c(1, 2, 3),
y = c("a", "b", "c"),
z = c(TRUE, FALSE, TRUE)
)tidyverse package, as tibble is from it.
tibble
There is another equivalent way of writing it rowwisely. For example,:
tribble(
~x, ~y,
1, "a",
2, "b",
3, "c"
)tibble).
This is perfect when typing in smaller datasets by hand.
Tibbles come with plenty of benefits over base R’s classic data frame—particularly in how they track and preserve data types. You’ll notice each column explicitly labeled as dbl (double) or chr (character), which can save you from those frustrating mysteries where your numeric data gets disguised as text.
Tibbles also enforce a neat and consistent data layout, called Tidy Data, which is a godsend when you’re dealing with other people’s messy spreadsheets. Quoting from Hadley Wickham (link):
Tidy datasets are easy to manipulate, model and visualize, and have a specific structure: each variable is a column, each observation is a row, and each type of observational unit is a table.

We will learn more about working with tibble more in the next sections, but for now, let’s move on to our example dataset.
Create a tibble with the following columns:
name with your name and surnameage with your ageis_student with a logical valuepenguinsWe’ll soon learn how to import data (another comedic ordeal in itself), but for now, let’s borrow the penguins dataset from the palmerpenguins package. It covers three penguin species—Adelie, Chinstrap, and Gentoo—and gives us some useful variables to play with. Who doesn’t love penguins?

First things first, let us load the penguins dataset.
# install.packages("palmerpenguins")
library(palmerpenguins)
data(package = "palmerpenguins")palmerpenguins package, you can install it by uncommenting this line.
palmerpenguins package.
penguins dataset.
As a first rule, always, always take a look at your data. The simplest way is simply to print it out:
penguinspenguins dataset.
This prints about the first couple rows of the dataset (another reason to love tibble over data frame in base R).
You may notice some NA values in the dataset. These are missing values, which are common in real-world datasets. We’ll learn how to handle them later.
If you’d like a better view of your dataset, try the skim() function from the skimr package:
pacman::p_load(skimr)
skim(penguins)skimr package.
penguins dataset.
| Name | penguins |
| Number of rows | 344 |
| Number of columns | 8 |
| _______________________ | |
| Column type frequency: | |
| factor | 3 |
| numeric | 5 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| species | 0 | 1.00 | FALSE | 3 | Ade: 152, Gen: 124, Chi: 68 |
| island | 0 | 1.00 | FALSE | 3 | Bis: 168, Dre: 124, Tor: 52 |
| sex | 11 | 0.97 | FALSE | 2 | mal: 168, fem: 165 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| bill_length_mm | 2 | 0.99 | 43.92 | 5.46 | 32.1 | 39.23 | 44.45 | 48.5 | 59.6 | ▃▇▇▆▁ |
| bill_depth_mm | 2 | 0.99 | 17.15 | 1.97 | 13.1 | 15.60 | 17.30 | 18.7 | 21.5 | ▅▅▇▇▂ |
| flipper_length_mm | 2 | 0.99 | 200.92 | 14.06 | 172.0 | 190.00 | 197.00 | 213.0 | 231.0 | ▂▇▃▅▂ |
| body_mass_g | 2 | 0.99 | 4201.75 | 801.95 | 2700.0 | 3550.00 | 4050.00 | 4750.0 | 6300.0 | ▃▇▆▃▂ |
| year | 0 | 1.00 | 2008.03 | 0.82 | 2007.0 | 2007.00 | 2008.00 | 2009.0 | 2009.0 | ▇▁▇▁▇ |
Think of skim() as your quick backstage pass, telling you how many rows, columns, missing values, and data types you’re dealing with. Trust me, a few seconds spent peeking at your data can save hours of confusion down the line.
iris is a widely-used dataset of plant traits. It is a default dataset in R. Take a look at the iris dataset using the skim() function.